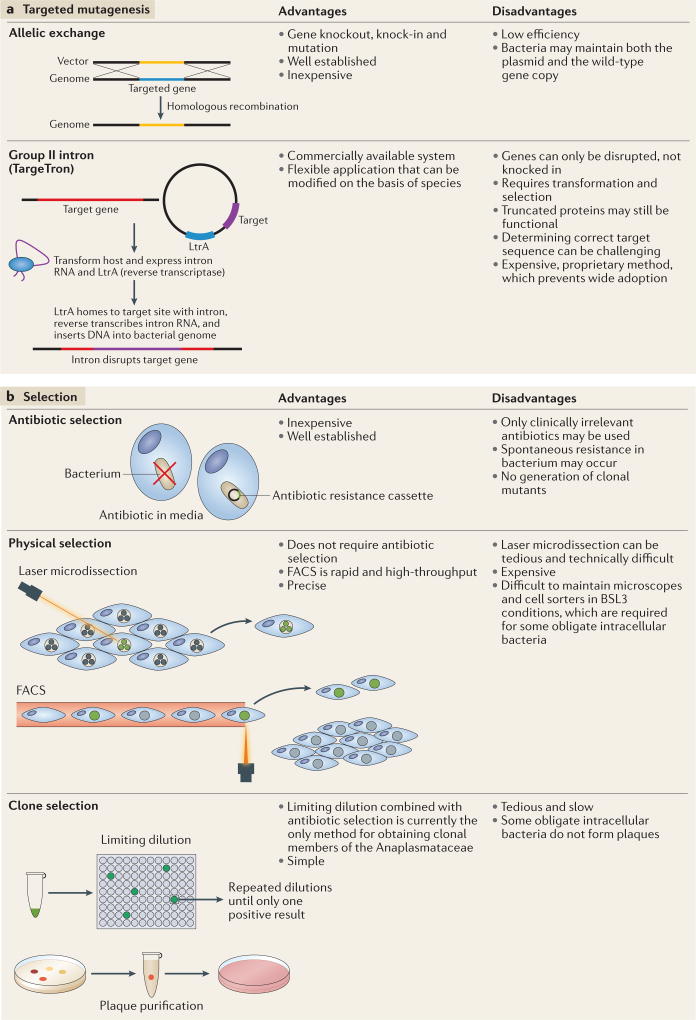
Figure 3. Targeted mutagenesis and selection.
a Targeted mutagenesis enables the alteration of specific bacterial genes. Allelic exchange can be used to introduce point mutations, and to insert and delete specific genes. Group II intron technology (TargeTron) enables introns to be specifically inserted into bacterial genes to disrupt gene function through LtrA, a multifunctional protein derived from Lactococcus lactis, which reverse transcribes and splices the intron and cleaves the recipient DNA for intron insertion51. TargeTrons have been successfully applied to generate transient mutants of Ehrlichia chaffeensis15 and stable mutants of Chlamydia trachomatis and Rickettsia rickettsii52,53. b | Two methods are currently used to distinguish mutants from wild-type bacteria: antibiotic selection and physical selection. Mutants can be physically separated from wild-type bacteria by fluorescence-activated cell sorting (FACS) and laser microdissection. One final step is obtaining clonal mutants, which can be isolated by limiting dilution, or, in some cases, plaque purification5. BSL3, biological safety level 3.