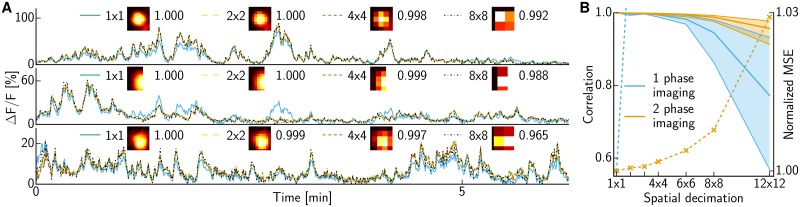
Fig 3. Low spatial resolution light-sheet imaging with previously identified cells.
(A) Inferred traces for the three neurons in Fig 2. Shapes were decimated by averaging 2×2, 4×4 or 8×8 pixels. The legend shows the resulting shapes Al as well as the correlation between the calcium traces Cl inferred from the decimated data versus the original traces C1 obtained from the non-decimated data. Further, the results obtained with merely 1-phase imaging for decimation by 8×8 pixels are shown as cyan traces. (B) Correlation (solid, left y-axis) between Cl and C1 for all cells recovered from this patch. Thick lines show the median, thin lines and shaded region the interquartile range (IQR). The correlation decays slowly as l increases for 2-phase imaging (orange) but drops abruptly if the shapes Al are estimated directly from decimated data (cyan) instead of being decimated from the shapes A1 estimated from a standard-resolution imaging phase. Mean-squared-error (dashed, right y-axis) normalized so that without decimation the value is 1.