Fig. 5.
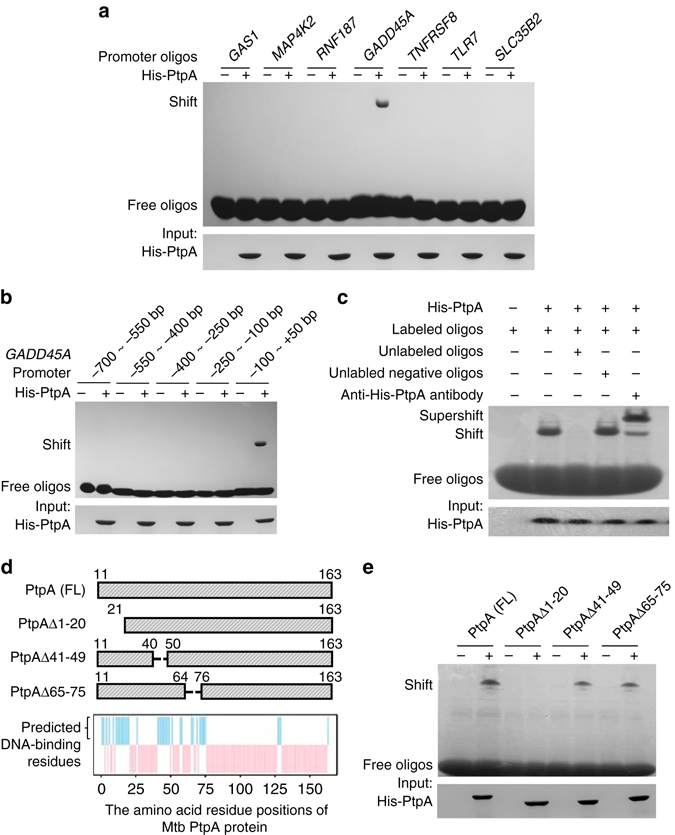
Identification of the DNA-binding region of potential target genes by PtpA. a EMSA analysis of interactions between His-tagged Mtb PtpA and its potential targeting gene promoters. The promoters of seven genes (10 nM) were amplified and incubated with 10 μg of purified His-PtpA. b EMSA analysis of interactions between His-tagged PtpA and five promoter fragments of GADD45A. The promoter fragments of GADD45A (10 nM) were amplified and incubated with 10 μg of purified His-PtpA. c Competitive EMSA analysis of specific interactions between PtpA and the promoter fragment of GADD45A (−100 bp to + 50 bp). Biotinylated oligos (−100 bp to + 50 bp) was used to determine their interactions with purified 10 μg His-tagged PtpA protein. Unlabeled positive (−100 bp to + 50 bp) and negative oligos (−250 bp ~ −100 bp) were added in lane 3 or 4 as specific competitor sequences. Lane 5 displayed a super-shift in the presence of anti-His-PtpA antibody. d The amino acid residue positions of Mtb PtpA potentially involved in interactions with DNA were predicted by DP-bind (blue bars). Top, schematic representation of PtpA deletion constructs used for EMSA analysis in e. FL, full-length. e EMSA analysis of interactions between Mtb PtpA proteins and its targeting promoter fragment (−100 bp to + 50 bp from the TSS of GADD45A). The biotinylated oligos were incubated with 10 μg of purified full-length or truncated Mtb PtpA proteins. Full blots are shown in Supplementary Fig. 10
