Figure 2.
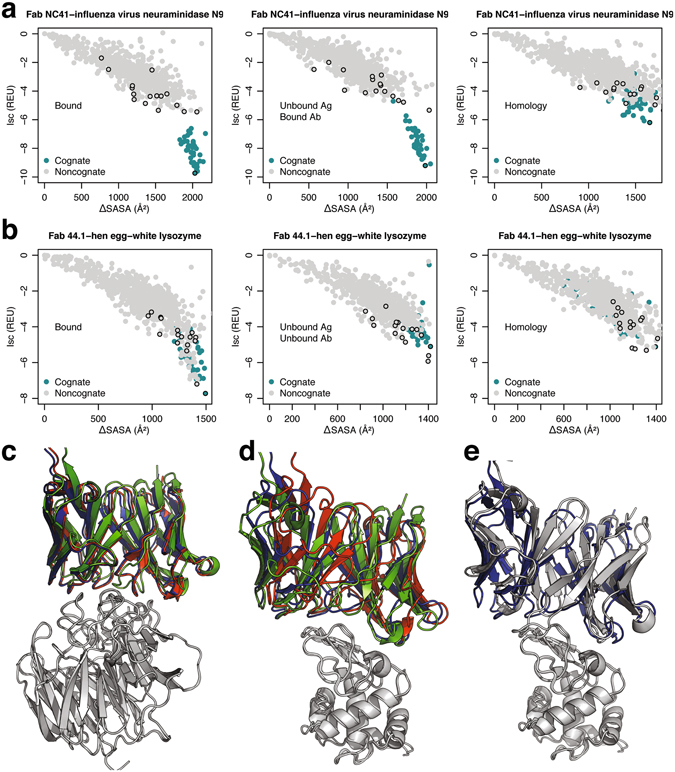
Binding discrimination in Fab NC41–influenza virus neuraminidase N9 and Fab 44.1–hen egg-white lysozyme complexes. Surface area change upon binding (ΔSASA) vs. Interface scores for model structures in antibody cross-docking tests using bound, unbound, and homology-modeled backbones in (a) influenza virus neuraminidase N9, and (b) hen egg-white lysozyme. Sixteen non-cognate structures (grey) are compared to cognate structures (turquoise); black outline indicates the top-scoring model for each antibody–antigen pair. The top-scoring cognate complexes generated using bound (red), unbound (blue), and homology (green) complexes in (c) influenza virus neuraminidase N9, and (d) hen egg-white lysozyme complexes. (e) Top-scoring cognate (blue) and non-cognate (grey) antibody against hen egg-white lysozyme.
