Fig. 4.
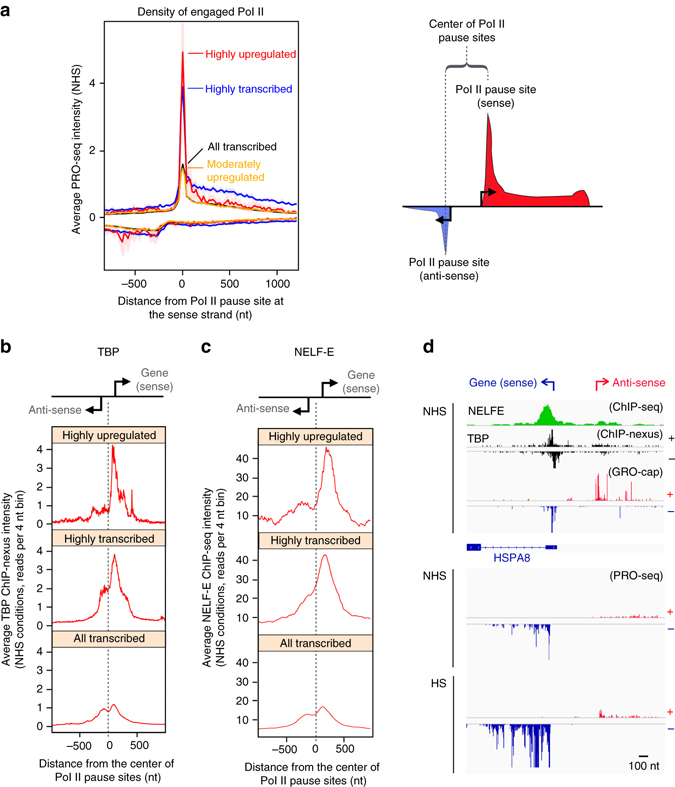
Directionality and rapid transcriptional induction is pre-wired in the promoter architecture. a PRO-seq profile of highly upregulated, highly transcribed, moderately upregulated and all transcribed genes, centered on the Pol II pause site at the coding strand. The intensity of transcriptionally engaged Pol II at the coding strand is indicated above the value zero, respective intensity at the non-coding strand is depicted with negative values. The right panel schematically depicts the Pol II profile at divergent regulatory elements, indicating TSSs to sense and anti-sense directions with arrows, Pol II pause sites at sense and anti-sense strands with dotted lines, and the mid coordinate between the Pol II pause sites with a bracket. b, c Average intensity of (b) TBP ChIP-nexus and (c) NELF-E ChIP-seq at highly upregulated, highly transcribed and all transcribed genes. The corresponding heatmaps are shown in Supplementary Fig. 4c. d Browser shot of HSPA8 gene indicating the positioning of NELF-E and TBP with respect to the GRO-cap mapping of transcription start sites at the sense and the anti-sense strands, and the PRO-seq profile of transcriptionally engaged Pol II prior to (NHS) and upon (HS) heat stress. ChIP-nexus data set for TBP was obtained from He et al.54, ChIP-seq for NELF-E is from the ENCODE55, and GRO-cap from Core et al.19
