Fig. 5.
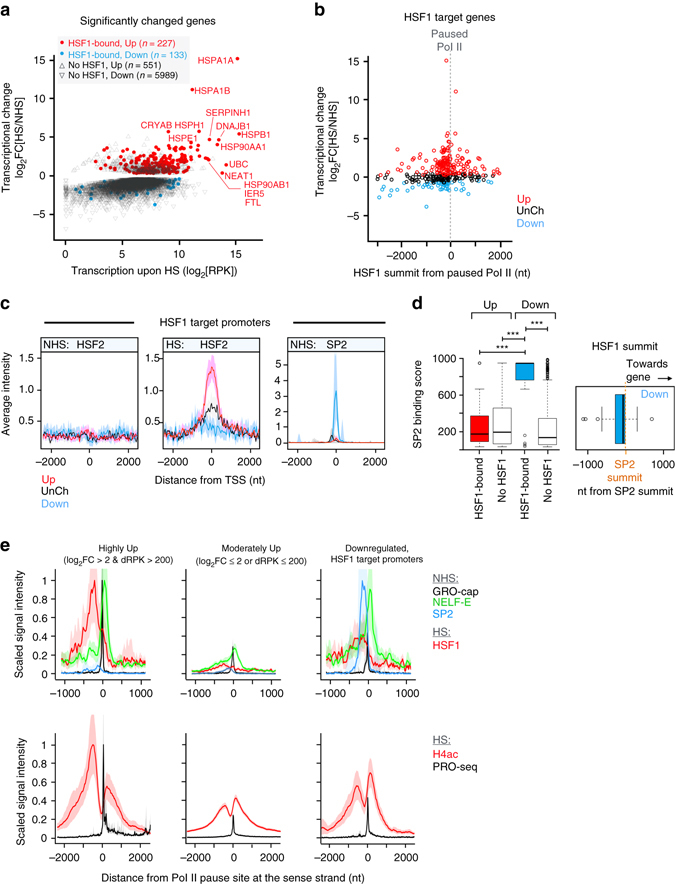
Local chromatin architecture is permissive or restrictive for HSF1-mediated trans-activation. a Transcriptional change of up and downregulated genes, plotted against the transcriptional level upon stress. The HSF1-bound upregulated genes are indicated with red, the HSF1-bound downregulated genes with blue closed circles. b Localization of HSF1 summit point from the Pol II pause site at the coding strand, plotted against the gene’s transcriptional change upon heat shock. c The average occupancy of HSF2 and SP2 at HSF1 target promoters. d Left panel: ENCODE binding score (proportional number between 0–1000; https://genome.ucsc.edu/FAQ/FAQformat.html#format12) of SP2 at the HSF1-bound and HSF1-unbound upregulated (Up) and downregulated (Down) genes. Significant P-values (Mann–Whitney U-test) are shown; the three asterisks indicating values lower than 0.0005. The position of HSF1 peak summit from the peak summit of SP2 is depicted with respect to the directionality of the divergent promoter (right panel). e Scaled ChIP-seq, PRO-seq and GRO-cap intensities at indicated gene groups. The highest average signal intensity for each factor in any bin across the gene groups is used as normalization constant, bringing the maximum signal to value 1. ChIP-seq data sets for NELF-E and SP2 were obtained from the ENCODE55, GRO-cap is from Core et al.19
