Extended Data Figure 2. Deletion of the distal enhancer in Hoxa11 intron using CRISPR-Cas9.
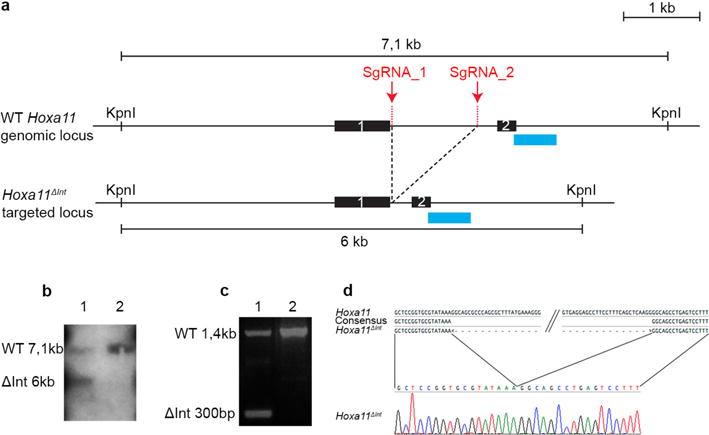
a, Scheme of the wild-type and targeted (Hoxa11ΔInt) loci. Sites targeted by the single-guide RNAS (sgRNA_1 and sgRNA_2) for the CRISPR-Cas9-mediated deletion of the distal enhancer. The blue rectangles indicate the position of the DNA probe used to confirm the deletion by Southern blot in b. b, Lane 1 shows the 6-kb KpnI band resulting from the CRISPR-Cas9-mediated deletion. Lane 2 was loaded with wild-type DNA. c, PCR reaction using a forward primer located upstream of sgRNA_1 and a reverse primer located downstream sgRNA_2 shows the presence of a 300 bp (ΔInt 300 bp) fragment expected for the Hoxa11ΔInt allele. d, The sequence of the 300-bp PCR fragment confirms the CRISPR-Cas9-mediated deletion of the Hoxa11 intronic region containing the distal enhancer (only the sequence encompassing the deletion breakpoints is shown).
