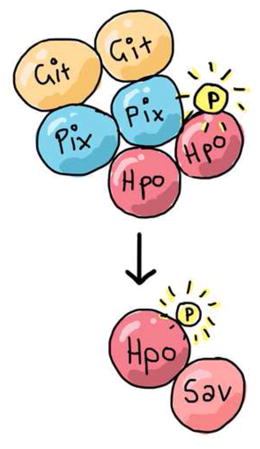
SUMMARY
The Salvador-Warts-Hippo (Hippo) pathway is a conserved regulator of organ size and is deregulated in human cancers [1]. In epithelial tissues, the Hippo pathway is regulated by fundamental cell biological properties, such as polarity and adhesion, and coordinates these with tissue growth [2–4]. Despite its importance in disease, development and regeneration, the complete set of proteins that regulate Hippo signalling remain undefined. To address this, we used proteomics to identify proteins that bind to the Hippo (Hpo) kinase. Prominent among these were PAK-interacting exchange factor (known as Pix or RtGEF) and G-protein-coupled receptor kinase interacting protein (Git). Pix is a conserved Rho-type guanine nucleotide exchange factor (Rho-GEF) homologous to Beta-PIX and Alpha-PIX in mammals. Git is the single D. melanogaster homologue of the mammalian GIT1 and GIT2 proteins, which were originally identified in the search for molecules that interact with G-Protein-coupled receptor kinases [5]. Pix and Git form an oligomeric scaffold to facilitate sterile 20-like kinase activation and have also been linked to GTPase regulation [5–8]. We show that Pix and Git regulate Hippo pathway-dependent tissue growth in Drosophila melanogaster, and that they do this in parallel to the known upstream regulator Fat cadherin. Pix and Git influence activity of the Hpo kinase by acting as a scaffold complex, rather than enzymes, and promote Hpo dimerization and autophosphorylation of Hpo’s activation loop. Therefore, we provide important new insights into an ancient signalling network that controls the growth of metazoan tissues.
Graphical Abstract
RESULTS and DISCUSSION
Pix and Git physically and genetically interact with Hippo
Hpo is recognised as the most upstream kinase in the Hippo pathway core kinase cassette [9–12]. Multiple proteins regulate its activity including Tao-1 [13, 14], RASSF [15] and the STRIPAK phosphatase complex [16]. To identify additional regulators of Hpo, we used affinity purification of Hpo tagged at both the N- and C-termini in D. melanogaster S2 cells, followed by mass spectrometry [17]. We recovered several known Hippo pathway proteins including Rassf, Salvador and Yki. Among the most abundant Hpo-interacting proteins were Pix and Git; Pix was identified with 5 peptides and a SAINT probability of 0.999, whilst Git was represented with 15 peptides and a maximal SAINT probability of 1 (Table S1) [18]. Furthermore, in an independent study, both Pix and Git were recovered as Hpo-binding proteins [16]. To confirm these results, we performed co-immunoprecipitation experiments in S2 cells and found that Hpo formed a physical complex with both Pix and Git (Figures 1A and S1). We also performed co-immunoprecipitation experiments in D. melanogaster wing imaginal discs and demonstrated that Pix and Git physically interact in this tissue (Figure 1B).
Figure 1. Pix and Git physically interact with Hippo and regulate Hippo pathway activity.
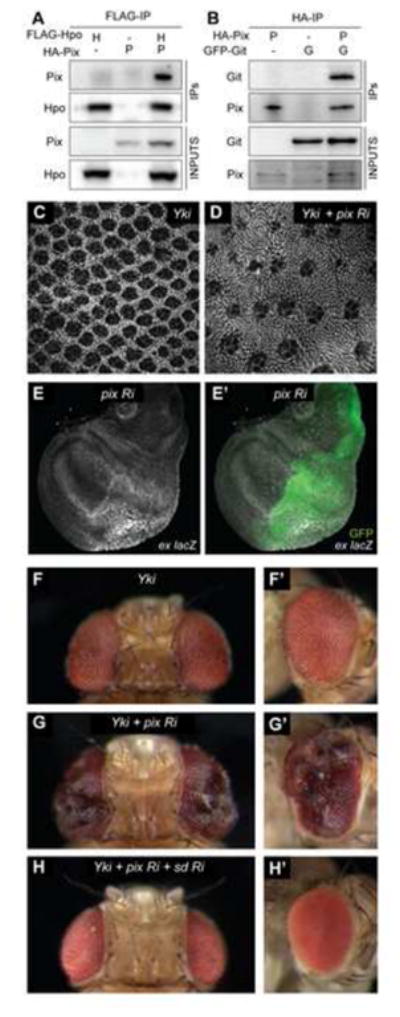
For co-immunoprecipitation experiments, protein lysates from S2 cells transfected with the indicated plasmids (A) or wing imaginal discs expressing the indicated transgenes (B) were incubated with anti-Flag or anti-HA antibodies, respectively. Western blot analysis was performed using anti-Flag, anti-HA and anti-GFP antibodies to reveal Hpo, Pix and Git, respectively. (C and D) D. melanogaster eyes 44 hours after puparium formation stained with anti-Discs-large. Eyes express UAS-Yki-S168A-YFP under the control of GMR-Gal4, either alone (C) or together with UAS-Pix RNAi (D). (E, E′) D. melanogaster third instar larval wing imaginal disc of the genotype: ex-lacZ, en-Gal4, UAS-GFP; UAS-Pix RNAi. Transcriptional activity of the ex gene was reported by β-galactosidase expression(grayscale) and in (E′) GFP (green) demarcates the posterior compartment, where Pix RNAi was expressed. (F–H′) Adult female D. melanogaster eyes, dorsal views in (F, G and H), lateral views in (F′, G′ and H′). Eyes express UAS-Yki-S168A-YFP under the control of GMR-Gal4, either alone (F, F′) with UAS-Pix RNAi (G, G′), or with UAS-Pix RNAi and UAS-Sd RNAi (H, H′). See also Figure S1.
Pix regulates Hippo pathway activity
To investigate a role for Pix in mediating control of organ size by the Hippo pathway, we performed a number of experiments. Initially, we depleted Pix using RNA interference (RNAi) in a sensitised Hippo pathway background (GMR>Yki) that we have used previously to identify novel Hippo pathway proteins [13, 19]. The GMR>Yki strain has eye specific Yki overexpression, which results in subtle eye overgrowth and an increase in interommatidial cells [13]. Pix depletion caused a striking increase in interommatidial cells in Yki-overexpressing eyes when assessed at 44 hours after puparium formation (APF) (Figures 1C and 1D). We also found in larval wing imaginal discs that Pix depletion caused an increase in both ex-lacZ and ban-lacZ, well-established reporters of Yki activity [20–22] (Figures 1E, 1E′, S1E and S1E′). The observed increase in ex-lacZ was dependent on Yki, as co-depletion of Pix and Yki suppressed this increase (Figures S1D and S1D′). Moreover, Pix depletion in GMR>Yki eyes gave rise to an increase in adult eye size, with folding and convolution of the eye, consistent with enhanced Yki activity (Figures 1F–1G′). Indeed, RNAi-mediated depletion of the key Yki transcription factor, Scalloped (Sd), completely suppressed the ability of Yki overexpression and Pix-RNAi to enhance eye size (Figures 1H and 1H′). Collectively, these in vivo data provide evidence that Pix regulates Hippo pathway dependent tissue growth.
Pix and Git limit tissue growth in parallel Fat cadherin
To assess the role of Pix and Git in tissue growth further, we assessed animals that were mutant for the genes encoding these proteins. Both pix and git mutant animals were semi-viable; adults that emerged displayed a crumpled wing phenotype, which precluded measurement of size (data not shown). No obvious overgrowth was observed in other adult tissues, such as the eye. A common theme with upstream regulators of the Hippo pathway is that they operate in a redundant fashion [23]. Double mutant analysis has revealed that several upstream Hippo pathway proteins can compensate for each other. For example the loss of either merlin (mer), fat, ex or kibra alone gives subtle overgrowth phenotypes whereas tissues that are double mutant for these sets of genes show very strong overgrowth [20, 24–27]. Therefore, we reasoned that Pix and Git’s ability to regulate Hippo pathway dependent tissue growth might be masked by functional redundancy. To test this idea, we made a series of double or triple mutant animals with either fat or ex (pix, fat; pix, fat, ex; and fat, git), as Fat and Ex are well-defined upstream Hippo pathway proteins [20, 28–31] and are thought to operate, at least in part, in separate branches of the Hippo pathway [23].
Initially we used the eyeless-FLP system to create clones of tissue lacking pix, fat, or both genes, in the eye and head capsule. Adult heads containing pix mutant clones displayed no obvious overgrowth whereas loss of fat showed slightly larger eyes and overgrowth in the head capsule, compared to controls (Figures 2A–2C). Compound mutation of both fat and pix, or fat and git genes caused substantial overgrowth, particularly in the ptilinum, compared to loss of each gene in isolation (Figures 2D and S2). These phenotypes were reminiscent of eye and head tissue that also lack both fat and ex, compared to tissue lacking only one of these genes (Figures 2C, 2E and 2F). Interestingly, tissue mutant for fat, pix and ex showed further overgrowth (Figure 2G). The enhanced tissue overgrowth in fat, pix and fat, ex, pix mutants was also manifest in reduced adult survival. Animals with fat, pix double mutant head tissue were less viable than animals with fat mutant head tissue alone, and animals with fat, ex, pix head tissue were 100% lethal (Figure 2H).
Figure 2. Pix and Git limit tissue growth in parallel to Fat cadherin.
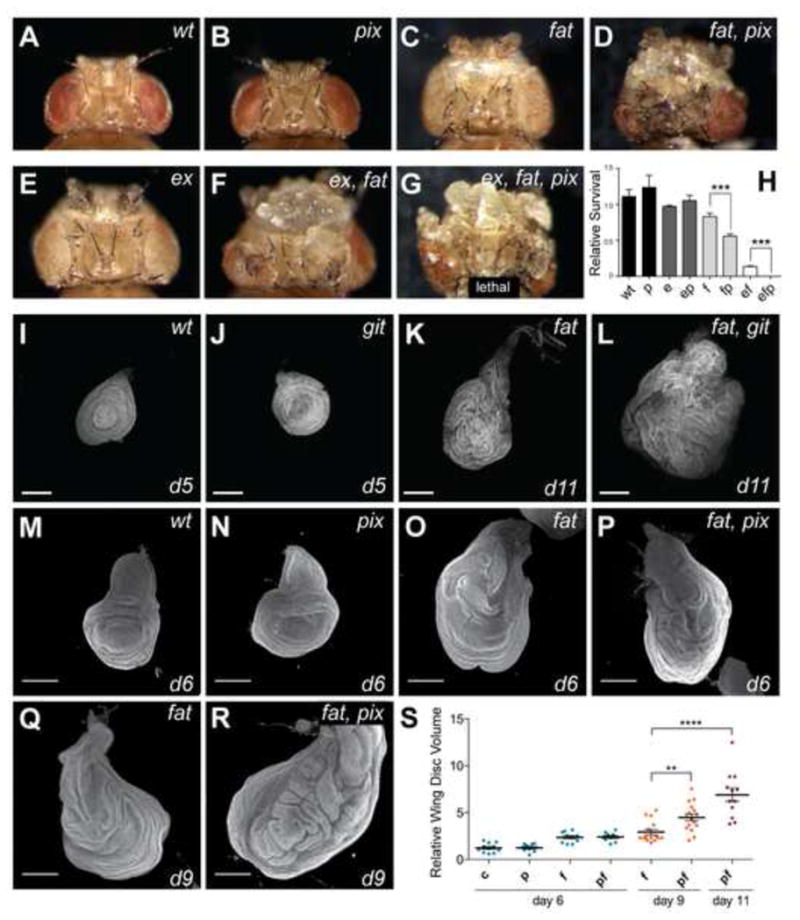
(A–G) Adult female D. melanogaster with head and eye tissue containing homozygous clones generated with eyeless-Flp of the following genotypes: A) wt; B) pix1036; C) fatfd; D) fatfd, pix1036; E) exe1; F) fat, ex; G) fatfd, exe1, pix1036.(H) Relative survival of the genotypes in (A–G) was determined by comparing the number of mutant progeny with the number of their wildtype siblings recovered from crosses. Genotypes of clonal tissue are: wt, wildtype P, pix; 1036 E, exe1; F, fatfd. At least 400 mutant or wildtype siblings were counted per genotype. Data represents mean +/− SEM. *** indicates a p-value <0.001. (I–R) Representative final size and age of third instar wing (I–L) and leg (M–R) imaginal discs for the indicated genotypes. Discs were stained with DAPI to mark nuclei. Scale bars = 150μM. (S) Quantification of wing disc volume of the indicated genotypes. c = control, f = fat, p = pix, fp = fat, pix. n=11, 10, 10, 10, 15, 16, 5 from left to right. data represents mean +/− SEM. **** indicates a p-value <0.0001, ** indicates a p-value <0.01. See also Figure S2.
Animals that were homozygous mutant for both fat and pix, or fat and git survived to early pupal stages of development, allowing assessment of organ size in larvae. We observed a striking increase in the size of imaginal tissues from fat, git compared to fat alone, which was especially obvious in leg discs at day 11 (Figures 2J–2L). We performed similar analyses on fat and pix single and double mutant wing discs, dissected from tightly developmentallystaged animals. Z -sections of non-flattened tissues were captured by confocal microscopy and the volume of each disc quantified (Figures 2M–2S). At day 6 pix mutant wing imaginal discs displayed no obvious increase in volume compared to wildtype wing discs. At day 9fat, pix discs were significantly larger than fat discs (Figure 2S). Because control, pix and git animals pupate at day 6 we were unable to quantify these tissues at day s9 or 11. In this experiment fat animal s pupated before day 11, fat, pix mutant animals did not, and contained even larger wing discs (Figure 2S). Together, these data support the idea that both Pix and Gitrestrict tissue growth, and do so in parallel to the Fat branch of the Hippo pathway.
Pix and Git control cell number through Hippo
Pix and Git are known to function together as a scaffold complex to activate the sterile 20-like kinase PAK [32], which is structurally related to Hpo. Given that we identified Pix and Git as Hpo-binding proteins, we investigated the possibility that these three proteins operate together to regulate tissue growth by analysing genetic interactions with hpo. We performed these experiments in pupal eyes, which offer an excellent setting to quantify increases in cell number when tissue growth is deregulated. We used the eyeless-FLP system to generate eyes with clones of tissue containing a hypomorphic allele of hpo (hpoMGH1). As reported previously, hpoMGH1 mutant eyes displayed an increase in the number of interommatidial cells compared to wildtype eyes (Figures 3A, 3B and 3H) [9]. We found that combining the hpoMGH1 allele with mutations in either git or pix further increased interommatidial cell number (Figures 3C, 3D and 3H). To test whether Pix and Git act separately or together to regulate the Hippo pathway, we analysed eye tissue that was triple mutant for pix, git and hpo. Triple mutant tissue showed no further increase in interommatidial cell number than either double mutant, suggesting that Pix and Git actin partnership to regulate the Hippo pathway (Figures 3E and 3H).
Figure 3. Pix and Git control cell number and organ size through Hippo.
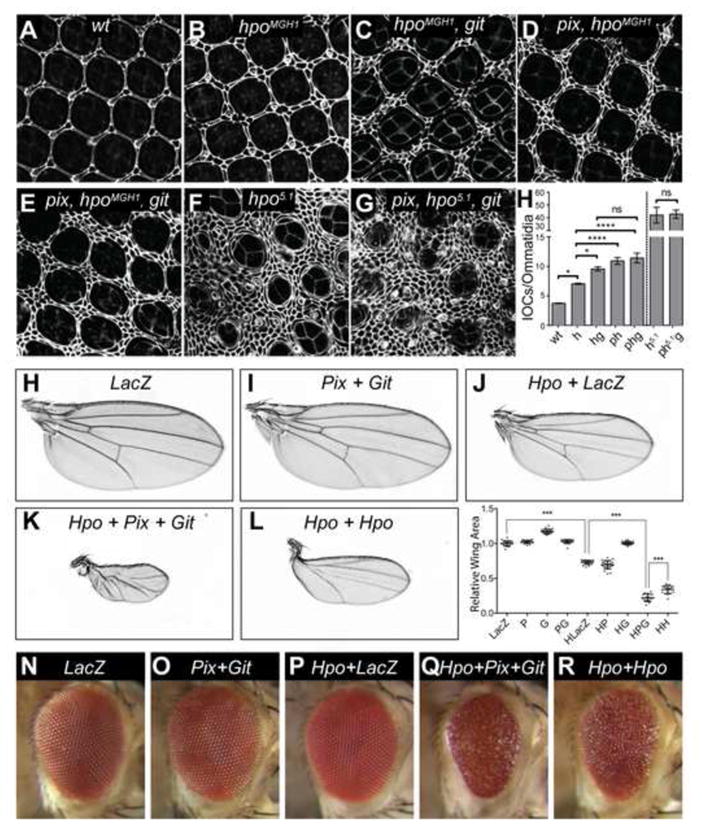
(A–G) D. melanogaster eyes (either wildtype or harboring the indicated mutations) 44 hours after puparium formation, stained with anti-Discs-large. (H) Quantification of interommatidial cell numbers of the genotypes displayed in (A–G). n=5 in (A), n=8 in (B), n=10 in (C), n=12 in (D), n=9 in (E), n=3 in (F), n=3 in (G). Data represents mean +/− SEM. * indicates a p-value <0.05, **** indicates a p-value <0.0001. Adult female D. melanogaster wings (H–L) and eyes (N–R) expressing the following transgenes under the control of nub-Gal4 or GMR-Gal4; (H) and (N) UAS-lacZ; (I) and (O) UAS-pix, UAS-git; (J) and (K) and (Q) UAS-hpo, UAS-pix, UAS-git;(L) and (R) UAS-hpo/UAS-hpo. (M) Quantification of wing area of the indicated genotypes. n=19 in (H), n=22 in (I), n=21 in (J), n=16 in (K), n=17 in (L). In (F), data represents mean +/− SEM. *** indicates a p-value <0.001. See also Figure S3.
To test whether Pix and Git regulate interommatidial cell number through Hpo or in parallel, we also examined their ability to affect cell number in tissue harbouring a null allele of hpo (hpo5.1), which lacks almost the entire Hpo coding sequence [33]. In the background of hpo nullizygous tissue, loss of pix and git no longer affected interommatidial cell number, suggesting that Pix and Git regulate cell number and eye growth by acting through Hpo (Figures 3F–3H).
Pix and Git function as a bipartite scaffold to promote Hippo dimerization and activity
Pix and Git perform many of their signal transduction functions by acting as a heterodimeric scaffold [34–37]. To test whether Pix and Git serve as a scaffold to activate Hpo, we used a series of genetic and biochemical experiments. Hippo pathway hyperactivation retards the growth of tissues such as the eye and wing [9–12, 38]. To determine whether Pix and Git could enhance Hpo’s ability to retard tissue growth, we used D. melanogaster strains harbouring pix and git transgenes. Pix and Git overexpression did not obviously affect wing or eye size when expressed using the nub-gal4 and GMR-Gal4 drivers, respectively, compared to controls (Figures 3H, 3I and 3M–3O). However, when we doubled the dosage of both pix and git transgenes, we observed a significant reduction in wing size, consistent with a role for Pix and Git in suppression of organ growth (Figure S3). Overexpression of a weak hpo transgene [39] reduced wing size to 60% of control wings, but had no obvious effect on eye size or roughness (Figures 3J, 3M and 3P). Individually expressing either Pix or Git with Hpo did not enhance Hpo’s ability to repress wing size (Figure 3M). However, when Pix, Git and Hpo were expressed together, a striking decrease in wing size was observed to only 15% the size of wildtype wings (Figures 3K and 3M). Consistently, co-expression of Pix, Git and Hpo in the eye substantially reduced eye size and increased roughness, compared to Hpo overexpression alone (Figure 3Q). In both eyes and wings, the observed enhancement of Hpo-driven inhibition of tissue growth by Pix and Git was greater than simply doubling the dose of the hpo transgene (Figures 3L, 3M and 3R). The observed requirement for both Pix and Git to be overexpressed simultaneously in order to enhance Hpo-driven growth retardation is consistent with the idea that Pix and Git function together in a complex to activate Hpo. The observation that Pix and Git overexpression minimally influence organ size, but enhance Hpo’s ability to limit organ size is reminiscent of experiments with Sav, which serves as a scaffold for Hpo and Wts; GMR-Gal4 dependent sav expression has no phenotype, but it enhances the ability of both Hpo and Wts to limit eye size [12, 38].
Hpo, and its mammalian orthologues, are known to dimerize and this is essential for kinase activation [39–41]. Based on our findings above, we predicted that Pix and Git would promote Hpo dimerization. To test this, we employed transgenic D. melanogaster strains expressing split Venus tagged-Hpo kinase-dead proteins HpoVC and HpoVN (wild-type Hpo was unsuitable as it caused substantial reduction of wing imaginal disc tissue when driven with nub-Gal4). In this system, Venus fluorescence is only detectable when Hpo dimerizes, and in doing so brings the N- and C-terminal halves of Venus together [39]. In control wing imaginal discs expressing HpoVC and HpoVN alone, as well as discs additionally expressing either Pix or Git, we observed very low Venus fluorescence (Figures 4A and 4C). When Pix and Git were expressed together, we observed a more than 3-fold increase in Venus fluorescence, indicative of strong induction of Hpo dimerization (Figures 4B and 4C). This effect was specific to Hpo dimerization, as Pix and Git overexpression failed to influence dimerization of the control proteins Fos and Jun (Figures 4C). Pix and Git overexpression also induced strong association of Hpo with Sav (Figure 4C), suggesting that Pix and Git influence Hpo’s ability to engage with and activate the Hippo pathway core kinase cassette, which is consistent with our findings that Pix and Git influence Yki activity and enhance Hpo’s ability to retard organ growth.
Figure 4. Pix and Git function as a bipartite scaffold to dimerize and activate Hippo.
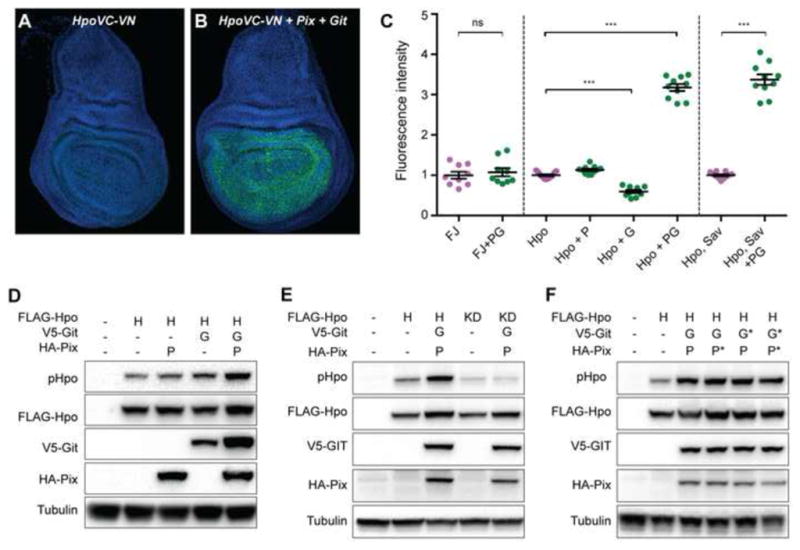
(A and B) Third instar larval wing imaginal discs stained with DAPI (blue). Both tissues express Hpo kinase-dead proteins fused to either the N- or C-terminus of Venus fluorescent protein (green). The tissue in (B) also expresses Pix and Git. (C) Quantification of Venus fluorescence in wing larval wing imaginal discs of the indicated genotypes (F=Fos, J=Jun, P=Pix, G=Git). n=9, 9, 11, 11, 10, 10, 11 and 10 from left to right. Data represents mean +/− SEM. *** indicates a p-value <0.001, whilst ns indicates no significant difference. (D–F) Western blot analysis of protein lysates from S2 cells transfected with the indicated plasmids. Hpo phosphorylation was detected using an antibody recognising phosphorylation of the Hpo activation loop (T195). Western blots were also probed with Anti-Flag, Anti-HA, Anti-V5 and Anti Tubulin to reveal total Hpo, Pix, Git and tubulin respectively. See also Figure S4.
To test biochemically whether Pix and Git activate Hpo, we assessed their ability to regulate phosphorylation of the Hpo activation loop (threonine 195), a well-characterized marker of activity of Hpo and its mammalian orthologues MST1 and MST2 [42, 43]. Overexpression of either Pix or Git alone in D. melanogaster S2 cells had no significant impact on Hpo activity, whereas Pix and Git co-overexpression resulted in an approximate doubling of Hpo T195 phosphorylation compared to Hpo alone (Figures 4D and S4F).
Pix and Git act as a scaffold, rather than enzymes, to activate Hippo
Given that Pix and Git promoted Hpo dimerization, we considered that they might activate Hpo by promoting Hpo transphosphorylation, a key mechanism of Hpo and MST1/2 activation in D. melanogaster and mammals, respectively [39, 41–43]. Alternatively, Pix and Git could modulate the activity of other kinases that are known to phosphorylate the activation loop of Hpo, such as Tao-1 [13, 14]. To distinguish between these two scenarios, we expressed either wildtype or kinase dead (KD, carrying a mutation in the ATP-binding site – K71R) versions of Hpo in the presence and absence of both Pix and Git, and assessed Hpo-T195 phosphorylation. Pix and Git only enhanced Hpo activation loop phosphorylation when coexpressed with active Hpo, suggesting that Pix and Git potentiate Hpo transphosphorylation rather than phosphorylation by additional regulatory proteins (Figure 4E).
Next, we more formally addressed whether Pix and Git act as scaffolds to activate Hpo as these proteins have been reported to function as both enzymes and scaffolds. For example, Pix enzymatic activity is required to activate the Rho-GTPases Rac1 and Cdc42, whereas Git enzymatic activity is required to deactivate ARF family GTPases. To discern between these possibilities, we generated mutant versions of Pix and Git that are known to abolish their enzymatic activity; Pix serine 89 was mutated to glutamic acid (S89E) [44], and Git arginine 39 was mutated to lysine (R39K) [32]. We then coexpressed wildtype or enzymatic dead (denoted by *) Pix and Git in the indicated combinations with Hpo (Figure 4F). In each scenario, Pix and Git overexpression were still able to robustly promote Hpo activation, as assessed by Hpo-T195 phosphorylation (Figure 4F). Given that in mammalian cultured cells, Pix and Git activate PAK [32], and PAK was recently linked to Hippo signalling [45], we considered the possibility that Pix and Git activate Hpo via the Drosophila homologues of PAK (Pak1 and Pak3), but found no evidence for this (Figure S4). Together, this indicates that Pix and Git activate Hpo by acting as scaffolds rather than enzymes, an assertion that is further supported by our gain of function data in cultured cells and in vivo, where overexpression of both Pix and Git were required to activate Hpo and enhance its ability to retard tissue growth. Pix and Git might enhance local concentrations of Hpo within cells to enable Hpo activation and/or facilitate structural changes that promote transphosphorylation between Hpo dimers. Such studies will be most informative in tissues that the Hippo pathway is known to control the growth of, such as imaginal discs.
CONCLUDING REMARKS
Most founding members of the Hippo pathway were identified in clonal homozygous screens. The present study highlights the importance of more recent biochemical and RNA interference approaches that have identified Hippo pathway genes that were not recovered using other methods. Further, this study underscores the high level of redundancy inherent in upstream regulators of the Hippo pathway, as growth regulatory roles for Pix and Git were only revealed when they were disabled in the context of mutations in the Fat upstream branch of the Hippo pathway. The Hippo pathway is known to regulate the growth of many tissues in addition to imaginal discs, and upstream regulators of the Hippo pathway show varying degrees of redundancy in these tissues [2, 4, 23]. Therefore, it is conceivable that Pix and Git regulate Hippo pathway activity in a non-redundant fashion in non-imaginal disc tissues. Currently it is unclear what controls Pix and Git in their ability to regulate Hpo. One possibility is that Pix and Git provide a link between the apicobasal polarity protein Scribble and the Hippo pathway. Scribble has been linked to Pix and Git in mammalian cells [35], and has been shown to affect Hippo pathway activity in both D. melanogaster and in mammalian cells [2, 4, 23]. A further intriguing possibility is that Pix and Git provide a connection between integrins, focal adhesions and Hippo signalling. Pix and Git are known to regulate focal adhesion turnover, and they localise at focal adhesions through Git’s ability to bind to Paxillin. Pix and Git are also well known mediators of mechanical information [46]. Therefore, in the context of tissue growth control, Pix and Git could conceivably provide a biochemical link between mechanical information from integrins and/or focal adhesions to the Hippo pathway core kinase cassette.
EXPERIMENTAL PROCEDURES
D. melanogaster stocks
Transgenic D. melanogaster stocks were generated that harbored the N-terminally tagged UAS-HA-Pix and UAS-MYC-Git coding sequences on the third chromosome. Other stocks were UAS-LacZ, UAS-GFP, GMR-Gal4, en-Gal4, nub-Gal4, y w eyFlp; FRT42D P[W+ ubi-GFP], UAS-Pix-1 RNAi (BSC#32974), ban-lacZ, and ex697(all Bloomington Drosophila Stock Center), pix (dpix1036) [47], git (dgitex21c) [48], hpoMGH1, hpo5.1, ft422, ftfd, exMGH1, exe1, UAS-Dicer, UAS-Pix RNAi (KK 105093), UAS-Yki RNAi (KK 104523), UAS-LacZ RNAi (GD 51446), UAS-Sd RNAi (KK 108877) UAS-Pak1 RNAi (KK 108937) and UAS-Pak3 RNAi (GD 39844) (all Vienna Drosophila RNAi Center). All D. melanogaster expressing transgenic constructs were reared at 25°C, with the exception of animals in Figures 3N–3R, which were raised at 18°C.
Immunofluorescence
Primary antibodies were specific for β-galactosidase (Sigma), Discs Large and Cubitus interruptus (both Developmental Studies Hybridoma Bank). Anti-mouse secondary antibodies were from Invitrogen. Tissues were stained as in [21]. DAPI was used to visualise nuclei in third instar wing and leg imaginal discs.
Quantification of organ size
Wings were dissected from adult female flies reared at 25°C, and mounted in Canada Balsam (Sigma). Wing sizes were quantified using Adobe Photoshop as in [21]. The mean and S.E.M. values of wing area were determined with GraphPad Prism. Flies laid eggs for four hours and wing imaginaldiscs were dissected from developmentally staged animals and stained with DAPI. Z-sections of non-flattened tissues were captured using a confocal microscope and Imaris software was used to quantify the volume of each disc. For statistical analysis, genotypes were compared using an analysis of variance (ANOVA)test, followed by a post hoc Tukey’s test to determine which genotypes were different from one another. When comparing only two genotypes, a Student’st test was used. p values <0.05 were considered significant.
Quantification of relative survival
FRT males that were heterozygous for a mutation of interest were mated to eyeless-FLP females. This mating is expected to produce F1 progeny that contain either wildtype or mutant clones at a one to one ratio. The ratio of wildtype and mutant F1 progeny were recorded for each genotype and compared. For statistical analysis, genotypes were compared using unpaired Student’s t tests. p values <0.05 were considered significant.
Expression plasmids
A complete Hpo open reading frame was cloned into pMK33-NTAP-GS or pMK33-CTAP-SG vectors [17], to generate N-or C -terminally tagged Hpo, respectively. D. melanogaster pix and git coding sequences in the pXJ40 plasmid were gift sfrom Dr E. Manser. To constitutively express these genes in cell culture, wesub -cloned N-terminally tagged HA-pix, and V5-gitsequences into the pAc5.1 vector. To generate transgenicD. melanogaster, N-terminally tagged HA-Pix and MYC -Git coding sequences were cloned into the pUAST vector. In order to express Pix protein without GEFenzymatic activity, we performed site directed mutagenesis to change serine 89 to glutamic acid (S89E) [44]. To express Git protein without GAP enzymatic activity, site directed mutagenesis was used to changearginine39 to lysine (R39K) [32]. pAc5.1-Hpo, pAc5.1-Hpo K71R and pAc5.1-RASSF were from N. Tapon [11, 15].
Affinity purification and mass-spectrometry
pMK33-TAP-GS Hpo plasmid constructs were used to establish stable S2 cell lines using hygromycin selection. Cells were induced with CuSO4 and protein extracts were prepared as in [17]. Extracts were incubated with streptavidin beads (Pierce), washed with lysis buffer, and proteins were eluted with 2 mM biotin in lysis buffer, precipitated with TCA, and separated on a short SDS-PAGE. Gel slices were submitted for mass spectrometry analysis and protein identification, which was performed at the Taplin Mass Spectrometry Facility at Harvard Medical School. Lists of identified proteins were statistically analyzed against six independent control samples from untransfected S2 cells using the SAINT program [18].
Immunoblotting
S2 cells were transfected with the indicated plasmids and lysed after 48 hours, whereas third instar larval imaginal discs were dissected and lysed directly. Lysates were immunoprecipitated, or directly subjected to SDS-PAGE and transferred to PVDF (Millipore). Membranes were immunoblotted with antibodies specific for HA tag (Invitrogen), Flag tag (Sigma), V5 tag (Invitrogen), GFP (Roche), phospho-T195-Hpo (Cell Signaling) or Tubulin (Sigma).
Supplementary Material
Data from experimental and control purifications were analyzed by SAINT. For each protein identified in the Hippo purification, the number of unique peptides is shown in column E (Spec). This number was compared to data from six biological replicates of control samples using extracts from untransfected S2 cells (column J, ctrlCounts). AvgP indicates the probability of a protein being a bona fide interactor, with values greater than 0.8 considered significant. Hippo itself is highlighted green (hpo), and Git and Pix (RtGEF) are highlighted yellow. Both Git and Pix were identified with a highly significant SAINT probability and were not detected in control purifications.
Supplemental Figure 1, related to Figure 1.
(A and B) Co-immunoprecipitation experiments were performed on protein lysates from S2 cells transfected with the indicated plasmids using anti-Flagantibodies to immunoprecipitate Hpo. In (A) Western blot analysis was performed using anti-Flag to reveal Hpo and anti-HA to reveal RASSF and Pix. Hpo immunoprecipitated with both RASSF and Pix. In (B) Western blot analysis was performed using anti-Flag to reveal Hpo, anti-HA to reveal Pix and anti-V5 to detect Git. Hpo immunoprecipitated with both Git, but only when Pix was co-expressed.
(C) The relative levels of pix mRNA to actin mRNA were assessed in third instar larval wing imaginal discsexpressing either lacZ RNAi or pix RNAi under the control of nubbin-Gal4. pix mRNA was significantly lower in tissues expressing pix RNAi. Data represents mean +/− SEM. * indicates a p-value <0.05, n=4.
(D–E′) D. melanogaster third instar larval wing imaginal discs expressing the indicated RNAi lines under the control of en -Gal4. Transcriptional activity of the ex gene (D and D′) or the ban gene (E and E′) were reported by β-galactosidase expression (grayscale). In the merged images GFP (green) demarcates the posterior compartment (D′), whereas in (E′) Cubitus interruptus staining demarcates the anterior compartment.
Supplemental Figure 2, related to Figure 2. Greater ptilinum overgrowth occurs when mutant for fat and git, compared to fat alone.
Adult female D. melanogaster heads from gitex21cmutant animals (A), containing homozygous fatfd clones generated with eyeless-Flp (B), or homozygous fatfd clones in a gitex21cmutant background (C).
Supplemental Figure 3, related to Figure 3. Pix and Git overexpression limits wing size.
Adult female D. melanogaster wings expressing the following transgenes under the control of nub-Gal4: (A) UAS-Dicer; (B) UAS-pix, UAS-git (two copies of each transgene and Gal4 driver). (C) Quantification of wing area of the indicated genotypes. n=15 in (A), n=16 in (B). Data represents mean +/− SEM. *** indicates a p-value <0.001.
Supplemental Figure 4, related to Figure 4. Pak1 and Pak3 depletion do not affect the ability of Pix and Git to enhance Hippo’s ability to retard wing size.
Adult female D. melanogaster wings expressing the following transgenes under the control of nub-Gal4: (A) UAS-lacZ; (B) UAS-hpo; (C) UAS-hpo, UAS-pix, UAS-git; UAS-lacZ RNAi; (D) UAS-hpo, UAS-pix, UAS-git; UAS-Pak1 RNAi; (E) UAS-hpo, UAS-pix, UAS-git; UAS-Pak3 RNAi. (F) Quantification of Hpo phosphorylation in (Figure 4D), n=3, error bars represent mean SEM. * indicates a p-value <0.05.
Acknowledgments
We thank S. Bahri, B. Baum, A.P. Haghighi, Z-C Lai, E. Manser, N. Tapon, the Developmental Studies Hybridoma Bank, the Vienna Drosophila RNAi Center, the Australian Drosophila Research Support Facility (www.ozdros.com), and the Bloomington Drosophila Stock Centre for fly stocks, plasmids and antibodies. We thank Liu Yang for generating SAINT scores. K.F.H is a Sylvia and Charles Viertel Senior Medical Research Fellow. This research was supported by a Project Grant from the National Health and Medical Research Council of Australia, and by NIH grant GM097727 and NSF grant 0640700 to A.V. Mass spectrometry was performed at the Taplin Mass Spectrometry Facility, Harvard Medical School.
Footnotes
AUTHOR CONTRIBUTIONS
L.G.D, C.L.C.P, X.Z., J.L.D, M.T., A.V. and K.F.H. performed experiments and analysed data. L.G.D and K.F.H conceived the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder MC, Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Seminars in cell & developmental biology. 2012;23:803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- 5.Premont RT, Claing A, Vitale N, Freeman JL, Pitcher JA, Patton WA, Moss J, Vaughan M, Lefkowitz RJ. beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci U S A. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 7.Feng Q, Albeck JG, Cerione RA, Yang W. Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. J Biol Chem. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- 8.Feng Q, Baird D, Cerione RA. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 2004;23:3492–3504. doi: 10.1038/sj.emboj.7600331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 10.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 11.Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 13.Poon CL, Lin JI, Zhang X, Harvey KF. The Sterile 20-like Kinase Tao-1 Controls Tissue Growth by Regulating the Salvador-Warts-Hippo Pathway. Dev Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 Phosphorylates Hippo/MST Kinases to Regulate the Hippo-Salvador-Warts Tumor Suppressor Pathway. Dev Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Kyriakakis P, Tipping M, Abed L, Veraksa A. Tandem affinity purification in Drosophila: the advantages of the GS-TAP system. Fly (Austin) 2008;2:229–235. doi: 10.4161/fly.6669. [DOI] [PubMed] [Google Scholar]
- 18.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nature methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poon CL, Zhang X, Lin JI, Manning SA, Harvey KF. Homeodomain-interacting protein kinase regulates hippo pathway-dependent tissue growth. Curr Biol. 2012;22:1587–1594. doi: 10.1016/j.cub.2012.06.075. [DOI] [PubMed] [Google Scholar]
- 20.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 21.Degoutin JL, Milton CC, Yu E, Tipping M, Bosveld F, Yang L, Bellaiche Y, Veraksa A, Harvey KF. Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat Cell Biol. 2013;15:1176–1185. doi: 10.1038/ncb2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herranz H, Hong X, Cohen SM. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr Biol. 2012;22:651–657. doi: 10.1016/j.cub.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 23.Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol. 2010;20:R574–582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW Domain Protein Kibra Acts Upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra Is a Regulator of the Salvador/Warts/Hippo Signaling Network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra Functions as a Tumor Suppressor Protein that Regulates Hippo Signaling in Conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 29.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 32.Loo TH, Ng YW, Lim L, Manser E. GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol Cell Biol. 2004;24:3849–3859. doi: 10.1128/MCB.24.9.3849-3859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genevet A, Polesello C, Blight K, Robertson F, Collinson LM, Pichaud F, Tapon N. The Hippo pathway regulates apical-domain size independently of its growth-control function. J Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–1011. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lecine P, Bellaiche Y, Dupont JL, Premont RT, Sempere C, Strub JM, et al. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 2004;14:987–995. doi: 10.1016/j.cub.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 36.Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank SR, Hansen SH. The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Seminars in cell & developmental biology. 2008;19:234–244. doi: 10.1016/j.semcdb.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 39.Deng Y, Matsui Y, Zhang Y, Lai ZC. Hippo activation through homodimerization and membrane association for growth inhibition and organ size control. Dev Biol. 2013;375:152–159. doi: 10.1016/j.ydbio.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Creasy CL, Ambrose DM, Chernoff J. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J Biol Chem. 1996;271:21049–21053. doi: 10.1074/jbc.271.35.21049. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, Zhang L. Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J Biol Chem. 2012;287:5784–5796. doi: 10.1074/jbc.M111.310334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Y, Pang A, Wang JH. Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis. J Biol Chem. 2003;278:11760–11767. doi: 10.1074/jbc.M211085200. [DOI] [PubMed] [Google Scholar]
- 43.Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- 44.Aghazadeh B, Zhu K, Kubiseski TJ, Liu GA, Pawson T, Zheng Y, Rosen MK. Structure and mutagenesis of the Dbl homology domain. Nature structural biology. 1998;5:1098–1107. doi: 10.1038/4209. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen HT, Hong X, Tan S, Chen Q, Chan L, Fivaz M, Cohen SM, Voorhoeve PM. Viral small T oncoproteins transform cells by alleviating hippo-pathway-mediated inhibition of the YAP proto-oncogene. Cell Rep. 2014;8:707–713. doi: 10.1016/j.celrep.2014.06.062. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 47.Parnas D, Haghighi AP, Fetter RD, Kim SW, Goodman CS. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron. 2001;32:415–424. doi: 10.1016/s0896-6273(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 48.Bahri SM, Choy JM, Manser E, Lim L, Yang X. The Drosophila homologue of Arf-GAP GIT1, dGIT, is required for proper muscle morphogenesis and guidance during embryogenesis. Dev Biol. 2009;325:15–23. doi: 10.1016/j.ydbio.2008.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data from experimental and control purifications were analyzed by SAINT. For each protein identified in the Hippo purification, the number of unique peptides is shown in column E (Spec). This number was compared to data from six biological replicates of control samples using extracts from untransfected S2 cells (column J, ctrlCounts). AvgP indicates the probability of a protein being a bona fide interactor, with values greater than 0.8 considered significant. Hippo itself is highlighted green (hpo), and Git and Pix (RtGEF) are highlighted yellow. Both Git and Pix were identified with a highly significant SAINT probability and were not detected in control purifications.
Supplemental Figure 1, related to Figure 1.
(A and B) Co-immunoprecipitation experiments were performed on protein lysates from S2 cells transfected with the indicated plasmids using anti-Flagantibodies to immunoprecipitate Hpo. In (A) Western blot analysis was performed using anti-Flag to reveal Hpo and anti-HA to reveal RASSF and Pix. Hpo immunoprecipitated with both RASSF and Pix. In (B) Western blot analysis was performed using anti-Flag to reveal Hpo, anti-HA to reveal Pix and anti-V5 to detect Git. Hpo immunoprecipitated with both Git, but only when Pix was co-expressed.
(C) The relative levels of pix mRNA to actin mRNA were assessed in third instar larval wing imaginal discsexpressing either lacZ RNAi or pix RNAi under the control of nubbin-Gal4. pix mRNA was significantly lower in tissues expressing pix RNAi. Data represents mean +/− SEM. * indicates a p-value <0.05, n=4.
(D–E′) D. melanogaster third instar larval wing imaginal discs expressing the indicated RNAi lines under the control of en -Gal4. Transcriptional activity of the ex gene (D and D′) or the ban gene (E and E′) were reported by β-galactosidase expression (grayscale). In the merged images GFP (green) demarcates the posterior compartment (D′), whereas in (E′) Cubitus interruptus staining demarcates the anterior compartment.
Supplemental Figure 2, related to Figure 2. Greater ptilinum overgrowth occurs when mutant for fat and git, compared to fat alone.
Adult female D. melanogaster heads from gitex21cmutant animals (A), containing homozygous fatfd clones generated with eyeless-Flp (B), or homozygous fatfd clones in a gitex21cmutant background (C).
Supplemental Figure 3, related to Figure 3. Pix and Git overexpression limits wing size.
Adult female D. melanogaster wings expressing the following transgenes under the control of nub-Gal4: (A) UAS-Dicer; (B) UAS-pix, UAS-git (two copies of each transgene and Gal4 driver). (C) Quantification of wing area of the indicated genotypes. n=15 in (A), n=16 in (B). Data represents mean +/− SEM. *** indicates a p-value <0.001.
Supplemental Figure 4, related to Figure 4. Pak1 and Pak3 depletion do not affect the ability of Pix and Git to enhance Hippo’s ability to retard wing size.
Adult female D. melanogaster wings expressing the following transgenes under the control of nub-Gal4: (A) UAS-lacZ; (B) UAS-hpo; (C) UAS-hpo, UAS-pix, UAS-git; UAS-lacZ RNAi; (D) UAS-hpo, UAS-pix, UAS-git; UAS-Pak1 RNAi; (E) UAS-hpo, UAS-pix, UAS-git; UAS-Pak3 RNAi. (F) Quantification of Hpo phosphorylation in (Figure 4D), n=3, error bars represent mean SEM. * indicates a p-value <0.05.


