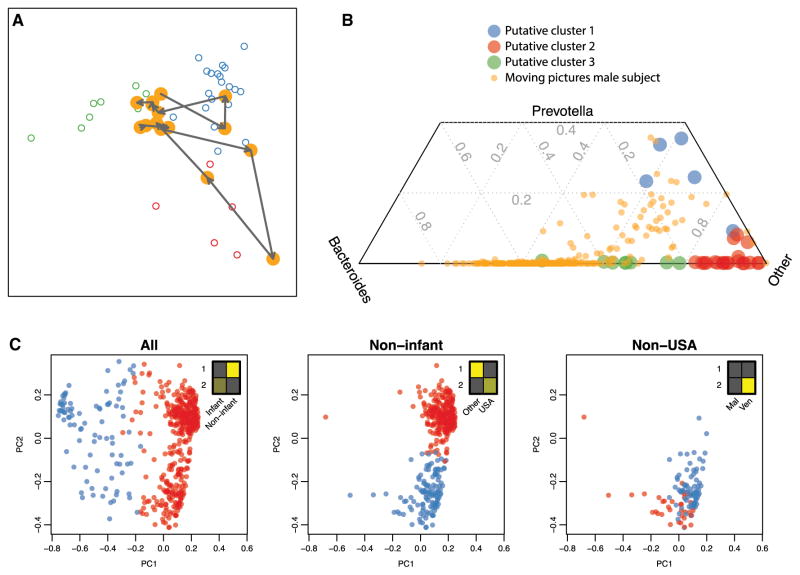
Figure 3. Enterotypes Can Be Unstable, Continuous, and Driven by Sampling Frame.
(A and B) Genus-level enterotype time series superimposed on putative clusters derived from 33 subjects (Arumugam et al., 2011). (A) Two selected trajectories of consecutive daily samples are shown for a single male subject (Caporaso et al., 2011). Meta-HIT samples are colored by putative enterotype cluster. The two selected trajectories show the subject’s microbiome profile “walking” from one putative enterotype to another over the course of several days. (B) Ternary plot of composition of Bacteroides, Prevotella, and other genera daily for a year for a single subject (Caporaso et al., 2011) and for published cross-sectional samples (Arumugam et al., 2011). These analyses demonstrate the temporal fluidity of enterotypes and provide clear proof by counterexample that enterotypes are not discrete states that separate individuals.
(C) Clustering performed on nested sampling frames from individuals of wide-ranging age in three different countries (Yatsunenko et al., 2012). Methods were described previously (Arumugam et al., 2011). Insets show relative sizes of sample subsets (columns) in putative clusters 1 and 2 (rows). Clustering of all individuals, those over age 2, and those over age 2 and not living in the USA identify clusters driven by age, USA versus non-USA citizenship, and Malawi versus Venezuela citizenship, respectively (chi-square test, p = 2.5 × 10−66, 2.5 × 10−63, 8.6 × 10−4, respectively), demonstrating that cluster comembership between samples is driven by sampling frame.