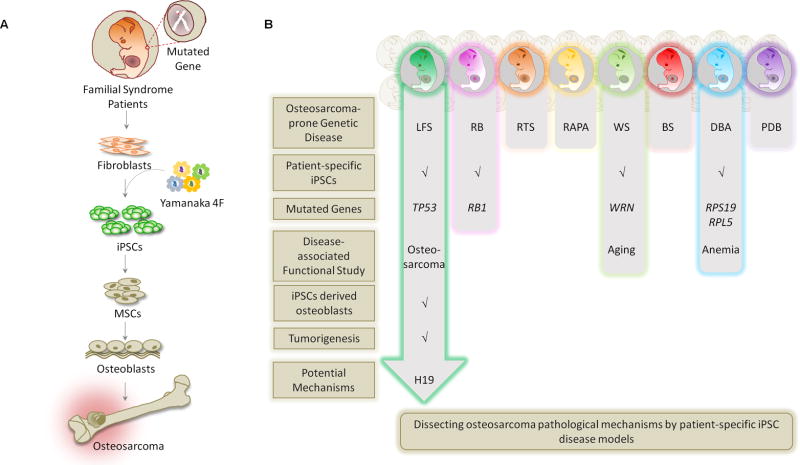
Figure 3. Patient-Derived iPSC Disease Models for Osteosarcoma.
(A) Patient-derived iPSCs are used to model human familial cancer syndromes and reveal a role of the mutant gene(s) in disease development. To apply iPSC methodology to study genetic disease-associated bone malignancy, patient fibroblasts are biopsied from skin and then reprogrammed to iPSCs by the four "Yamanaka factors" (OCT4, SOX2, KLF4 and c-MYC). iPSCs are then differentiated into MSCs, and then further, to osteoblasts. These iPSC-derived osteoblasts can be examined for osteoblast differentiation defects and tumorigenic ability. Systematic comparison of the genome/transcriptome/interactome between mutant and wild-type osteoblasts can further elucidate pathological mechanisms. (B) Current progress of applying LFS, RB, RTS, RAPA, WS, BS, DBA and PDB patient-derived iPSCs to model disease etiology and dissect disease-associated osteosarcomas.