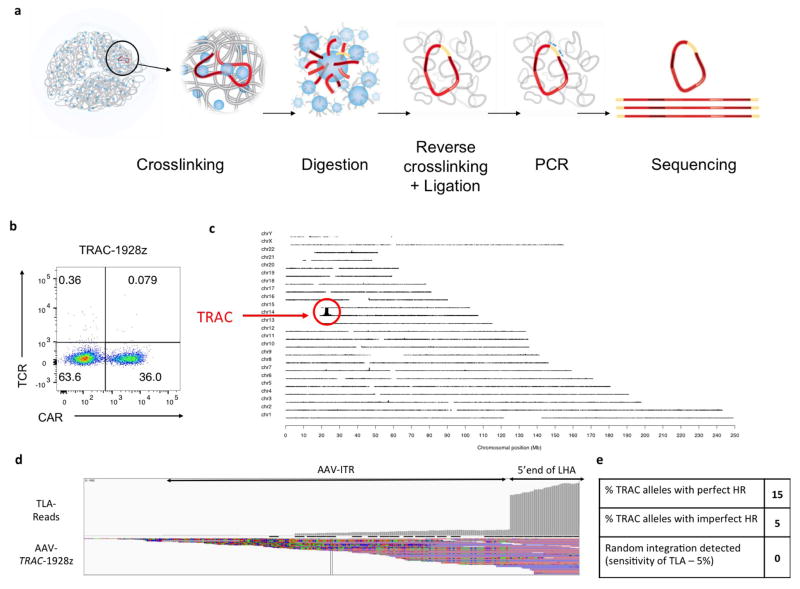
Extended Data Figure 2. Whole-genome mapping of the AAV6 TRAC-1928z integration using the TLA technology.
a, Schematic representation of the TLA technology17. For this study, two sets of primers targeting the CAR and the left homology arm have been used. b, TCR/CAR FACS plot of the TRAC-1928z CAR T cells used for the TLA analysis. CAR T cells have been processed as in Extended Data Fig. 1b and expanded for 2 weeks. c, TLA sequence coverage across the human genome using 1928z CAR specific primers (CD28-specific forward: 5′-ACAATGAGAAGAGCAATGGA-3′ and scFV-specific reverse: 5′-GAGATTGTCCTGGTTTCTGT-3′). The chromosomes are indicated on the y axis, the chromosomal position on the x axis. TRAC-encoded CAR T cells were produced as in Fig. 1 and expanded for 10 days before processed for analysis. The primer set was used in an individual TLA amplification. PCR products were purified and library prepped using the Illumina NexteraXT protocol and sequenced on an Illumina Miseq sequencer. Reads were mapped using BWA-SW, which is a Smith–Waterman alignment tool. This allows partial mapping, which is optimally suited for identifying break-spanning reads. The human genome version hg19 was used for mapping. d, TLA sequence coverage aligned on the AAV-TRAC-1928z sequence (Targeting sequence flanked by ITRs). The grey vertical bars on top represent the coverage at the shown positions. The coverage showed integration of the AAV ITRs in fraction of reads. The coverage comparison between ITR and CAR integration at the 5′ and 3′ ends of the TRAC homology arms locus allow the measurement of faithful and unfaithful homologous recombination shown in e. e, Final results from the TLA analysis.