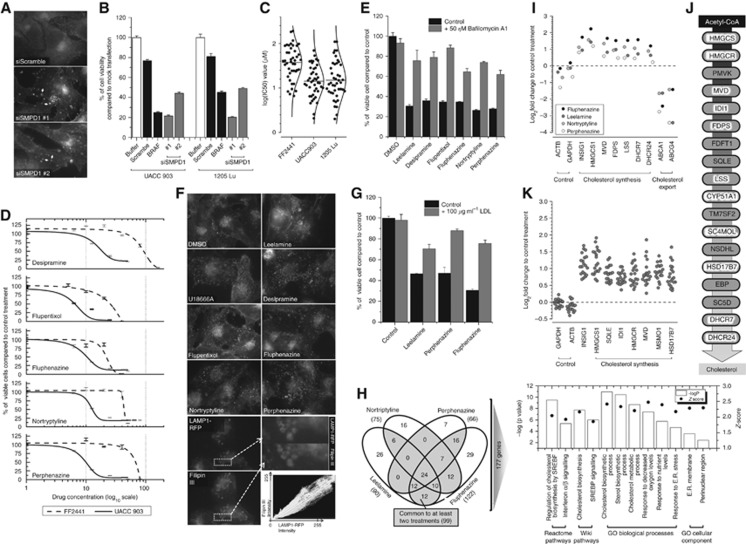
Figure 1.
ASM inhibitors induce melanoma-specific cell death by inhibiting intracellular cholesterol transport. (A) fluorescence microscopy of intracellular cholesterol localisation following knockdown of SMPD1 (ASM1) as detected by Filipin-III staining (arrows show accumulated cholesterol); (B) the viability of UACC 903 and 1205 Lu melanoma cells following knockdown of SMPD1; (C) IC50 values of various melanoma cell lines and FF2441 fibroblasts treated with ASM inhibitors. Dose–response curves were drawn in OriginPro (OriginLab) using Levenberg Marquardt algorithm; (D) viability of UACC 903 and FF2441 cells following 24 h of treatment with increasing concentrations of ASM inhibitors; (E) viability of melanoma cells treated with various ASM inhibitors in the absence or presence of v-ATPase inhibitor, Bafilomycin-A1; (F) intracellular cholesterol localisation following leelamine, U18666A or ASM inhibitor treatments as detected by Filipin-III staining. (lower) Co-localisation of RFP-tagged lysosomal LAMP1 protein with cholesterol; (G) LDL treatment protects UACC 903 cells from ASM inhibitor-mediated cell death; (H) Venn diagram showing number of significantly altered genes identified by RNA-sequencing of UACC 903 cells treated with ASM inhibitors (left); and enrichment analysis of significantly altered 177 genes; (I) expression level of cholesterol synthesis genes following ASM treatment as detected by RNA-sequencing; (J) cholesterol synthesis pathway. White highlighted genes were identified as significantly deregulated by RNA-sequencing; (K) distribution of log-2-fold change in expression levels of cholesterol synthesis genes identified by microarray analysis following treatment of MCF7 cells with ASM inhibitors.