Abstract
Archaeal fibrillarin (aFib) is a well-characterized S-adenosyl methionine (SAM)-dependent RNA 2′-O-methyltransferase that is known to act in a large C/D ribonucleoprotein (RNP) complex together with Nop5 and L7Ae proteins and a box C/D guide RNA. In the reaction, the guide RNA serves to direct the methylation reaction to a specific site in tRNA or rRNA by sequence complementarity. Here we show that a Pyrococcus abyssi aFib–Nop5 heterodimer can alone perform SAM-dependent 2′-O-methylation of 16S and 23S ribosomal RNAs in vitro independently of L7Ae and C/D guide RNAs. Using tritium-labeling, mass spectrometry, and reverse transcription analysis, we identified three in vitro 2′-O-methylated positions in the 16S rRNA of P. abyssi, positions lying outside of previously reported pyrococcal C/D RNP methylation sites. This newly discovered stand-alone activity of aFib–Nop5 may provide an example of an ancestral activity retained in enzymes that were recruited to larger complexes during evolution.
Keywords: fibrillarin, Nop5, C/D RNP, RNA 2′-O-methylation, Archaea
INTRODUCTION
Abundant noncoding RNAs, such as rRNAs, tRNAs, and snRNAs, have a large variety of modified nucleotides important for RNA folding, stability, and function (Helm 2006) (http://mods.rna.albany.edu/mods/) (Limbach et al. 1994). Among universal RNA modifications, ribose 2′-O-methylation is particularly abundant in rRNAs and tRNAs in all three domains of life and is believed to be among the most ancient RNA modifications (Poole et al. 2000; Grosjean 2005). The 2′-O-methyl group favors a C3′-endo conformation of the ribose, which enhances local stacking and rigidity, confines hydrogen bond formation, and increases hydrophobic surface. In addition, it protects against alkaline hydrolysis and nuclease attack (Decatur and Fournier 2002; Motorin and Helm 2011). Evidence has been found that archaea growing at elevated temperatures have an increased number of 2′-O-methylated RNA nucleotides to support RNA folding and stabilization (Noon et al. 1998; Dennis et al. 2001). Although individual modified nucleotides in rRNA are not highly conserved and are often dispensable, they all cluster at functionally important sites, and their global depletion results in severe growth defects (Decatur and Fournier 2002; for review, see Yip et al. 2013).
In Eukarya and Archaea, most of the 2′-O-methylriboses are produced by an RNA-dependent mechanism, so-called C/D ribonucleoprotein (RNP) particles (for review, see Yip et al. 2013). Similarly with the widely described CRISPR systems that attack specific sequences in DNA, the target specificity of the box C/D RNP modification reaction is determined by sequence complementarity of the antisense elements to their RNA targets (Kiss-László et al. 1996). Archaeal C/D RNPs comprise one box C/D guide RNA for two protein sets composed of L7Ae, Nop5, and aFib (the methyltransferase—aFib in Archaea; Fibrillarin or Nop1p in Eukarya) proteins. The C/D guide RNAs are typically 50–70 nt long and contain conserved sequence motifs named C (RUGAUGA) and D (CUGA) boxes at their 5′- and 3′-ends, respectively, as well as internal C′ and D′ boxes. These motifs fold into Kink-turn (K-turn) (Klein et al. 2001) and Kink-loop (K-loop) (Nolivos et al. 2005) structures, respectively, which serve as a platform for the C/D RNP assembly (Tripp et al. 2016). In a hierarchical manner, L7Ae has been shown to bind a K-structure in C/D guide RNA, followed by recruitment of a preassembled aFib–Nop5 heterocomplex (Bortolin et al. 2003; Tran et al. 2003; Gagnon et al. 2012). Finally, self-association of Nop5, observed in crystals and in solution, drives the formation of a higher order C/D RNP comprising two copies of C/D guide RNAs and four copies of each protein (Aittaleb et al. 2003; Oruganti et al. 2007; Bleichert et al. 2009; Lapinaite et al. 2013). A hallmark feature of the box C/D guide RNA is its sequence complementarity with target RNA. Located immediately upstream of the D or D′ box, a predefined sequence of 10–21 nt, termed antisense element, directs the 2′-O-methylation onto the target nucleotide in substrate RNA paired to the fifth nucleotide upstream of the D or D′ box sequence (Cavaillé et al. 1996; Kiss-László et al. 1996; Nicoloso et al. 1996).
However, the proposed structural model could not explain certain reported observations pointing at additional important interactions between the RNP components. At high concentrations, aFib–Nop5 complexes have been shown to bind the C′/D′ motif in guide RNA (Tran et al. 2003) and in pre-tRNA-Trp guide-substrate (Bortolin et al. 2003) in the absence of L7Ae; in the latter case, substantial methylation activity of the C/D pre-tRNP complex has also been observed (Bortolin et al. 2003), suggesting that aFib–Nop5 can utilize a substrate-borne C/D guide RNA independently of L7Ae. UV-crosslinking and RNA fragment binding studies revealed that interactions of Nop5 with internal loops outside the C/D box are important for proper complex formation (Ghalei et al. 2010; Bower-Phipps et al. 2012).
In this work, we examined the capacity of aFib to act independently of C/D guide RNA. We found that Pyrococcus abyssi aFib–Nop5 heterodimer performs targeted 2′-O-methylation of ribosomal 16S and 23S rRNA in the absence of C/D RNA guide and L7Ae in vitro. The mapped methylation sites in 16S rRNA (G47, G516, and C847, or 41, 550, and 876 in Escherichia coli numbering, respectively) are located at or close to the central core of the small ribosomal subunit. Our results suggest that the aFib–Nop5 heterodimer can function autonomously in the formation of 2′-O-methylribose without engaging the C/D RNA guidance system.
RESULTS
aFib–Nop5 heterodimer methylates 16S and 23S rRNA independently of C/D guide RNA
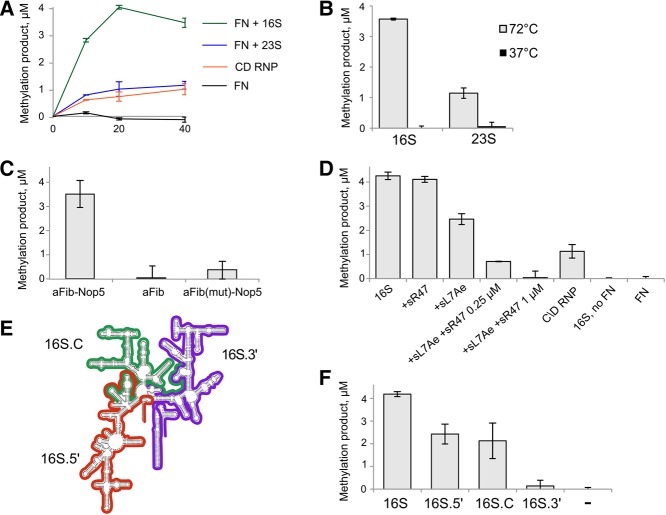
To test the RNA guide-independent activity of an aFib–Nop5 heterodimer, recombinant aFib and Nop5 proteins from the hyperthermophilic archaeon P. abyssi were coexpressed in E. coli and copurified to homogeneity by affinity chromatography. The purified aFib–Nop5 heterodimer was incubated with a series of RNA substrates at 72°C under conditions previously described for the reconstituted P. abyssi C/D sR47 guide RNP particle (Tomkuvienė et al. 2012). As shown in Figure 1, we observed aFib–Nop5-dependent incorporation of radiolabeled methyl groups in the case of in vitro transcribed P. abyssi 16S and 23S rRNA substrates; however, no methylation was detectable with P. abyssi 5S rRNA, tRNA-Leu(CAA), Mus musculus tRNA-Asp, polyU, and sonicated herring sperm DNA (Supplemental Fig. S1A). The observed activity was comparable to or higher than that of a fully assembled C/D sR47 RNP complex which targets C34 in the tRNA-Leu(CAA) transcript (Fig. 1A; Nolivos et al. 2005). Note that the bona fide reaction was saturated at equimolar amounts of C/D sR47 RNP and tRNA-Leu substrate leading to one methyl group incorporated per molecule of substrate. Unexpectedly, we observed a total of 4 µM methylation products per 1 µM 16S rRNA substrate after incubation with 1 µM aFib–Nop5 (Fig. 1). This strongly suggested that more than one site within the 16S rRNA substrate was methylated.
FIGURE 1.
P. abyssi aFib–Nop5 heterodimer methylates P. abyssi 16S and 23S rRNA. Reactions containing 1 µM aFib–Nop5 (FN), 100 µM [methyl-3H]-SAM, and 1 µM in vitro transcribed substrate RNAs (as indicated) were incubated at 72 °C for 40 min. C/D RNP reactions contained 1 µM C/D RNP, assembled with sR47 guide RNA, and substrate tRNA-Leu(CAA). (A) [methyl-3H]-incorporation time courses of the full C/D RNP and aFib–Nop5 with 16S or 23S rRNAs. (B) Temperature dependence of the aFib–Nop5 methylation activity. (C) Methylation activity of a separate aFib protein and aFib–Nop5 and mutant aFib(D150A)–Nop5 heterodimers. (D) aFib–Nop5 activity on 16S rRNA is inhibited by the assembly of a full C/D RNP. aFib–Nop5 and 16S rRNA reaction was supplemented with 3 µM L7Ae and either 0.25 µM or 0.5 µM sR47 C/D guide RNA. Reactions with only one additional C/D RNP component contained either 3 µM L7Ae or 1 µM sR47. (E) Three truncated P. abyssi 16S rRNA substrates: 16S.5′—red, 16S.C—green, and 16S.3′—purple. Secondary structure map according to Cannone et al. (2002). (F) aFib–Nop5 activity on truncated 16S rRNA substrates 16S.5′, 16S.C, and 16S.3′.
To exclude the possibility of exogenous 2′-O-methylation activity contamination, identical reactions were performed at 37°C. As for the C/D guide sR47 reaction (Supplemental Fig. S1B) and other C/D RNP reactions from thermophilic archaea (Omer et al. 2002; Hardin and Batey 2006), the aFib–Nop5 heterodimer activity was observed at 72°C but not at 37°C (Fig. 1B and Supplemental Fig. S1B), as expected for a thermophilic enzyme (Vieille and Zeikus 2001).
We also tested if aFib alone could perform the 2′-O-methylation reaction. Although a separately purified aFib protein was able to act in the context of the reconstituted C/D sR47 RNP particle, attesting to its full functional integrity (data not shown), we observed no incorporation of methyl groups to 16S rRNA with aFib alone under similar conditions (Fig. 1C).
Based on previous studies of a C/D RNP from Archaeoglobus fulgidus, which identified conserved residues important for cofactor binding and catalytic activity (Aittaleb et al. 2004), we produced and examined in our system the aFib D150A mutant (corresponding to aFib D133A in the above study). We found that this mutation leads to a nearly complete loss of methylation (Fig. 1C) confirming a functional importance of this residue for the activity of aFib–Nop5. This finding also suggests that the same cofactor binding pocket is operational in both the aFib–Nop5 heterodimer and in the native C/D RNP.
The stand-alone aFib–Nop5 activity is inhibited by the assembly of a full C/D RNP particle
We further tested whether the observed aFib–Nop5 activity toward 16S rRNA is affected by the presence of L7Ae and sR47 C/D guide RNA (sR47 lacks target sites on 16S rRNA). We observed partial inhibition of the reaction, containing 1 µM aFib–Nop5, with 0.25 µM of the C/D guide sR47 RNA-L7Ae, and complete inhibition in the presence of 0.5 µM sR47-L7Ae (equal to 1 µM aFib–Nop5 binding sites). Addition of equimolar amounts of sR47 guide RNA or L7Ae had no or partial inhibitory effect, respectively (Fig. 1D). We conclude that the assembly of a fully functional C/D sR47 RNP abolishes the aFib–Nop5 stand-alone methylation reaction, and addition of the other RNP components sequesters aFib–Nop5 to different degrees from acting on its stand-alone 16S rRNA substrate. This result is consistent with previous data showing that aFib–Nop5 heterodimer binds to a preformed C/D guide sRNA-L7Ae complex but not the C/D guide sRNA by itself (Bortolin et al. 2003).
The specificity of the aFib–Nop5 stand-alone methylation activity
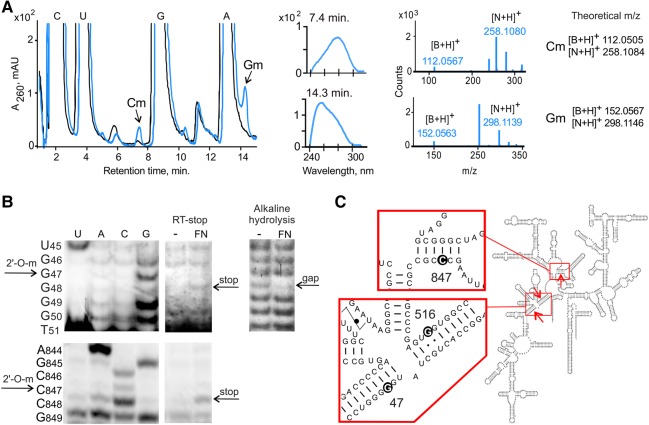
To investigate the specificity of the aFib–Nop5 heterodimer stand-alone activity, we performed in vitro modification of several types of RNA substrates corresponding to the whole or different domains of the P. abyssi 16S rRNA, referred to here as 16S.5′, 16S.C, and 16S.3′ (Fig. 1E). We found that the aFib–Nop5 complex was able to modify 16S, 16S.5′, and 16S.C RNA substrates but not 16S.3′ RNA (Fig. 1F). As expected, the methylation was only observed when the reactions were performed at 72°C (Supplemental Fig. S1C). However, we could not detect any methylation activity using shorter fragments of P. abyssi 16S RNA (corresponding to 431–533, 856–962, and 1320–1512 positions) which have been predicted to contain a high density of 2′-O-modifications in vivo (Gaspin et al. 2000; Omer et al. 2000; Dennis et al. 2001; McCloskey and Rozenski 2005) (data not shown). To precisely locate the methylation sites in the 16S.5′ and 16S.C RNAs, we performed HPLC/MS analysis of nuclease P1 or RNase A or RNase T1 digestion products. Table 1 summarizes the identified methylated species; Figure 2A and Supplemental Figure S2A show corresponding HPLC/MS data. For the 16S.5′ and 16S.C RNA substrates, two (GUGmGU/GGmG) and one (GCCmCG) consensus products containing 2′-O-methyl groups were identified after nuclease treatment, respectively. Since the GUGGU sequence is unique in 16S.5′ RNA, we can conclude that the G516 position (550 E. coli numbering) is 2′-O-methylated. In the other cases, there were 16 occurrences of GGG and four occurrences of GCCCG in 16S.5′ and 16S.C RNA primary sequence, respectively. To further locate the modified nucleotides, we performed RNase H digestion of the modified 16S.5′ and 16S.C RNA products. The digestion was directed by 18–20-mer DNA oligonucleotides that hybridized in between the predicted methylation sites. Subsequently, gel-purified RNA fragments were digested with nuclease P1 and analyzed by HPLC/MS. This approach permitted unequivocal allocation of 2′-O-methyl groups to positions G516 and C847 (876 E. coli numbering) in 16S.5′ and 16S.C RNAs, respectively (Supplemental Fig. S2B). The other 2′-O-methylated site in 16S.5′ was identified within a GGGGG pentanucleotide located at positions 46–50 (Supplemental Fig. S2B). To determine which of the Gs was modified, we performed a primer extension assay on an RNaseH-derived shorter fragment (positions 1–93) of 16S.5′ RNA and identified a reverse transcriptase (RT) arrest at position G47 (41 E. coli numbering); this finding was further confirmed by alkaline hydrolysis (Fig. 2B, upper panel). In the same manner, we observed an RT arrest at position C847 in the full-length 16S rRNA preincubated with aFib–Nop5 (Fig. 2B, lower panel).
TABLE 1.
Identification of methylated nucleotides in aFib–Nop5-modified 16S rRNA substrates using nuclease digestion and HPLC-MS analysis
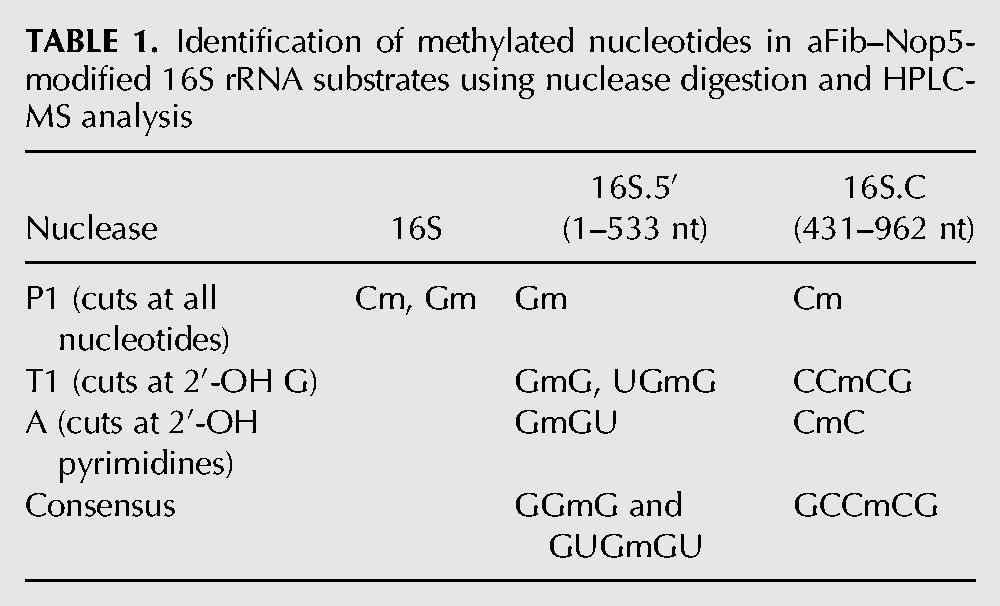
FIGURE 2.
Identification of methylation products by HPLC/MS and reverse transcription analysis. (A) HPLC/MS analysis of P1 nuclease-digested 16S rRNA substrate after incubation with aFib–Nop5 (blue) and control in the absence of aFib–Nop5 (black). UV chromatograms are shown together with UV absorption and mass spectra (Hall 1971) of the indicated peaks. N denotes nucleoside; B denotes nucleobase. (B) Sequence mapping of aFib–Nop5 modified sites. (Upper panels) analysis of modified 16S.5′ (subfragment 1–93) using an RT-stop assay (left) and alkaline hydrolysis with a subsequent primer extension (right). (Lower panel) analysis of modified 16S rRNA using an RT-stop assay. 2′-O-modified nucleotides correspond to enhanced bands (RT-stop) or gaps (alkaline hydrolysis) in the sequencing ladders. (C) Schematic depiction of aFib–Nop5 target sites G47, G516, and C847 in P. abyssi 16S rRNA.
Altogether, we found that aFib–Nop5 alone confers SAM-dependent 2′-O-methylation of G47, G516, and C847 positions in 16S rRNA transcript in vitro (Fig. 2C). Similar analysis showed no methylation of these positions in 16S rRNA isolated from cultivated P. abyssi cells (Supplemental Fig. S5).
Structural determinants of the stand-alone methylation specificity
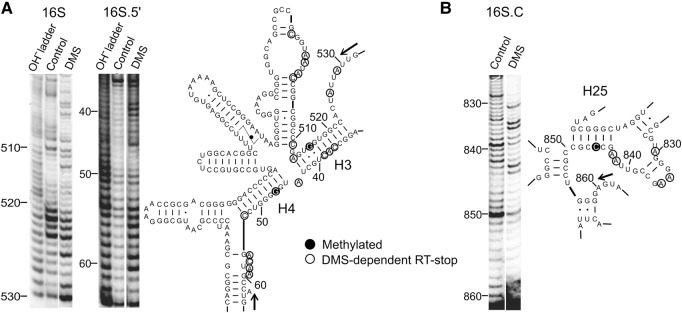
The identified three methylation sites showed no apparent sequence similarity or discernible unique features within the secondary structure of P. abyssi 16S rRNA (Fig. 2C). Interestingly, G516 methylation was observed only in the context of 16S and 16S.5′ but not in 16S.C or a short RNA fragment (positions 431–533) pointing at specific structural requirements for the stand-alone activity. Productive reaction complexes of bona fide C/D RNPs require full complementarity of the 10-mer guide RNA–target RNA heteroduplex (Yang et al. 2016), and even single mismatches in the heteroduplex have been shown to reduce or completely abolish the methylation efficacy of aFib (Singh et al. 2004; Appel and Maxwell 2007). In the predicted 16S rRNA secondary structure map (Cannone et al. 2002), all three target positions are located in double-stranded regions (helices H3, H4, and H25) (Fig. 3). We therefore performed dimethyl sulfate (DMS) footprinting experiments to probe the secondary structure of substrate 16S.5′ and 16S.C RNAs in the regions concerned. Consistent with similar studies on the whole small subunit rRNA (Ding et al. 2014), our results showed that H25 was completely protected from the DMS attack, whereas H3 and H4 showed substantial reactivity of certain A and C nucleotides, indicating a lower stability of the latter two helices when compared with H25 (Fig. 3). This observation indicates that the stand-alone activity is quite tolerant to variations in helix stability. Interestingly, a eukaryal fibrillarin not only acts in C/D RNP complexes to methylate RNA, but also has recently been shown to methylate the H2A histone (Tessarz et al. 2014; Loza-Muller et al. 2015), which reflects the ability of this enzyme class to embrace different substrates in the active site.
FIGURE 3.
DMS footprinting analysis of RNA secondary structure of aFib–Nop5 target regions of 16S and 16S.5′ RNA (A) and 16S.C (B) (Left panels) probed by DMS. (Right panels) secondary structure models of the full-length P. abyssi 16S rRNA (only areas of interest are shown) (Cannone et al. 2002). Full circles mark aFib–Nop5 methylation targets, open circles mark nucleotides that induce reverse transcription stops after DMS treatment, that is, unpaired cytosines and adenines. Arrows denote the start and direction of the reverse transcription primer extension. Control—no DMS.
Three-dimensional structure of the substrate is certainly an important element in directing aFib–Nop5 to its specific nucleotide targets. To this end, we tested the roles of two known K-turn motifs in 16S rRNA (Wimberly et al. 2000; Schroeder et al. 2010) in directing the aFib–Nop5-dependent methylation. We therefore replaced Kt-11 or Kt-23 in H11 (5′ domain) or H23 (central domain), respectively, by simple Watson–Crick double-stranded regions (Supplemental Fig. S3). In both cases, we observed aFib–Nop5-dependent methylation of the 16S.5′ and 16S.C substrates (data not shown). Thus, the K-structures do not appear to be important for the stand-alone activity of aFib–Nop5, in contrast to their essential roles in bona fide archaeal C/D RNPs (Omer et al. 2006; Tripp et al. 2016). Further, we inspected the 3D structure of the bacterial ribosome small subunit (Wimberly et al. 2000) and found that two target sites (C847 and G47) are located on one face of the RNA molecule separated by 70 Å from each other (Supplemental Fig. S4). Given the span (∼80 Å) of the two active sites (Aittaleb et al. 2003) and the described conformational flexibility of the aFib–Nop5 heterotetramer (Oruganti et al. 2007), it appears likely that these two sites can be reached from a single-binding position on the 16S rRNA. The third methylated site (G516) is fairly close to G47, but appears to be better approachable from the opposite face of the ribosomal subunit. The latter reaction apparently requires a different binding position of aFib–Nop5, although a single-face-approach scenario for the modification of all three sites cannot be excluded in light of possible changes in rRNA structure in solution when compared with the crystal derived model.
DISCUSSION
The present study for the first time demonstrates the capacity of the aFib–Nop5 heterodimer, a component of C/D box RNP from P. abyssi, to perform targeted 2′-O-methylation of pyrococcal 16S rRNA transcript in vitro independently of C/D guide RNA and the RNA binding protein L7Ae. Altogether, this finding is in line with previous reports showing that aFib and Nop5 form a precomplex before the assembly of the C/D guide RNP particle (Aittaleb et al. 2003; Bortolin et al. 2003; Tran et al. 2003; Gagnon et al. 2012), Nop5 can interact directly with the RNA substrate (Hardin et al. 2009) and the requirement of Nop5 for the binding of aFib on the RNA substrate (Omer et al. 2002). In addition, Nop5 conserved residues have been shown to facilitate the binding of SAM cofactor in the active site of aFib (Aittaleb et al. 2004) and to be important for aFib activity via an undetermined mechanism (Gagnon et al. 2012).
We precisely mapped the methylated residues at positions G47, G516, and C847 within the 16S rRNA (Fig. 2C). The specificity determinants of this “unguided” enzymatic reaction remain largely unknown, as no common sequence or structural features among the target sites could be identified. On the other hand, the target residues are adjacent to the functional decoding center of the small ribosomal subunit (Wimberly et al. 2000). This region has recently been defined as a core domain that acts as a hub, linking the four peripheral domains and imposing their positioning and orientation (Gulen et al. 2016). In addition, this region also exhibits the highest density of predicted C/D RNP-directed 2′-O-methylation target sites (Dennis et al. 2015), which presumably contribute to the stabilization of the core domain and the decoding center. An equivalent position to G47 has been predicted to be a target for the C/D RNP-directed methylation in Methanopyrus kandleri (Dennis et al. 2015). Even though G516 and C847 are not predicted targets of C/D RNP, some of their neighboring nucleotides are (Gaspin et al. 2000; Dennis et al. 2015). Therefore, it is likely that the observed catalytic capacity of the aFib–Nop5 heterocomplex is a relic of an ancestor 2′-O-methylation of 16S rRNA in primordial cells.
Contrary to our expectations, we could not detect 2′-O-methylation at the in vitro target positions in rRNA isolated from cultured P. abyssi cells. Instead, we found the presence of 2′-O-methyl groups at neighboring nucleotides (Supplemental Fig. S5), which is consistent with predicted C/D RNP methylation targets in P. abyssi (Gaspin et al. 2000; Dennis et al. 2015). As C/D RNAs have been found to be abundant in archaeal cells (Dennis et al. 2015), it is likely that the aFib–Nop5 heterodimer produced in P. abyssi cells is fully recruited into C/D RNPs, and thus none or little of it remains available to confer detectable levels of the stand-alone activity (see above). Interestingly, some mesophilic archaea, like Halobacterium salinarum, have been reported to have little if any detectable C/D RNAs but still have retained the Nop5–aFib operon (Dennis et al. 2001), and there are reported cases of C/D RNAs guiding tRNA methylation in Haloarchaea (Clouet d'Orval et al. 2001; Joardar et al. 2011). But the pattern of ribosomal RNA modifications in Haloarcula marismortui has been shown to resemble a bacterial pattern rather than that of a thermophilic archaea (Hansen et al. 2002). Thus we scanned the genomic sequences of seven representatives of Halobacteriacea for possible C/D RNAs targeting their 16S rRNAs using SnoScan (Lowe and Eddy 1999), and found only a few low significance hits (Supplemental Table S1). This result may argue for unique features of halobacterial C/D guide RNAs (Joardar et al. 2011) or point at potential stand-alone methylation activity of the aFib–Nop5 proteins in the cells, which remains to be experimentally tested.
The self-capacity of aFib–Nop5p to carry out ribose methylation could be related to the observed activity of the pseudouridine (Ψ) synthase Cbf5 outside of the H/ACA RNP context. Archaeal H/ACA RNPs and C/D RNPs share common features in addition to guiding RNA modification by base-pairing (Kiss-László et al. 1996; Baker et al. 2005). Both types of RNP particles comprise specific guide RNAs containing Kink RNA structures (K-loop and/or K-turn) which are specific binding sites for the ribosomal L7Ae protein (Rozhdestvensky et al. 2003). In addition to Cbf5 and L7Ae, archaeal H/ACA RNP also contains Nop10p and Gar1 proteins (for review, see Yip et al. 2013). Interestingly, the archaeal Cbf5 pseudouridine synthase can also perform uridine isomerization in an RNA-independent, or stand-alone, mechanism on ACA-less-tRNA substrates at position U55, and U2603 of 23S rRNA (Roovers et al. 2006; Muller et al. 2007, 2008; Zhou et al. 2011). Furthermore, Cbf5 has been shown to preferentially bind the H/ACA box RNA over its stand-alone substrate, resulting in similar inhibition of the stand-alone activity (Zhou et al. 2011). Both C/D and H/ACA RNPs are believed to have evolved from a primitive translation apparatus (Tran et al. 2004), or rRNA-processing RNAs (Lafontaine and Tollervey 1998). The finding that archaeal Cbf5 may function in protein-only as well as RNA-guided manner suggested that Cbf5 could be a direct descendant of a primordial Ψ synthase (Roovers et al. 2006). Indeed, Cbf5 is homologous to bacterial tRNA:Ψ55 pseudouridine synthase TruB (Koonin 1996). It is proposed that gene duplication of the TruB-like enzyme in Eukaryotic lineage gave rise to Ψ synthases Pus4p that retained the original specificity for tRNA, and Cbf5 that became available for involvement in an RNP function (Lafontaine and Tollervey 1998). Similarly, aFib is homologous to RrmJ stand-alone 2′-O-methyltransferases acting on 23S rRNA in Bacteria to Eukarya, but the duplication giving rise to the RrmJ and the fibrillarin lineages predates the LUCA (Feder et al. 2003). aFib evolved tightly associated with Nop5. This is reflected by the fact that they are associated in the same operon in archaeal genomes. Interestingly, eukaryal fibrillarin has been shown to have retained/regained the ability to bind RNA nonspecifically (Rakitina et al. 2011). Moreover, other archaeal methyltransferases acting on RNA have been shown to possess double activities shared by two enzymes in further evolutionary lineages (Urbonavičius et al. 2014).
Altogether these findings provide convincing arguments that both archaeal RNA-modifying enzymes, aFib and Cbf5, retained the capacity to act independently of corresponding guide RNP particles reflecting their common history and providing a rear glimpse of an intermediate state in the ladder of evolutionary genesis of complex multicomponent biological systems.
MATERIALS AND METHODS
Expression and purification of proteins
The P. abyssi recombinant proteins were expressed from three derived pET15b vectors: two of them had L7Ae or aFib genes cloned separately (Bortolin et al. 2003), whereas the third had Nop5 and aFib genes cloned together as a natural tandem, resulting in amino-terminally His-tagged L7Ae, aFib, and Nop5, respectively. The aFib D150A mutation was introduced into the aFib–Nop5 coexpression vector by the “megaprimer” PCR method (Sarkar and Sommer, 1990). The proteins were expressed in E. coli BL21 (DE3) RIL (Stratagene) cells after induction with 1 mM isopropyl-β-D-thiogalactoside. Following sonication, E. coli proteins were precipitated by thermodenaturation at 65°C for 15 min. Nucleic acids from aFib–Nop5 preparation were removed by precipitation with 2 M LiCl (1 h on ice), and from L7Ae preparation by addition of 0.15% polyethylenimine. His6-tagged proteins were purified by Ni2+ chelate chromatography on a HiTrap IMAC HP column (GE Healthcare). The heterodimer aFib–Nop5 was obtained by copurification using His6-tagged Nop5. After purification, the His6-tags were removed by thrombin (Amersham) treatment. L7Ae protein was further purified on a HiTrap SP-sepharose column (GE Healthcare).
RNA synthesis and purification
P. abyssi 16S and 23S rRNA genes were amplified by PCR from P. abyssi genomic DNA, using the Long PCR Enzyme Mix (Thermo Fisher Scientific), and primers (Metabion) 16S-5′Fw (TAATACGACTCACTATAGGGCGTACTCCCTTAATTCCGGTTGATCC, T7 RNA polymerase promoter is underlined), 16S-3′Rv (GATAGGAGGTGATCGAGCCGTAG) and 23S-5′Fw (TAATACGACTCACTATAGGGCTAAGCCGCCCGGTG), 23S-3′Rv (ACAGGACCTCGGGCGAT), respectively. 23S rDNA synthesis reaction contained 4% DMSO, and the ratio of 5′ and 3′ primers was 1:4. 16S rDNA was further used as a template for PCR amplification of fragment 431–533 nt, 856–962 nt, 1320–1512 nt, 16S.5′, 16S.C, and 16.S3′ DNA, using High Fidelity PCR Enzyme Mix (Thermo Fisher Scientific), and primers 16SI Fw (TAATACGACTCACTATAGGGCTTTTCCGGAGTGTAAAAAGCTCC) and 16SI Rv (CCAATAATAGTGGCCACCACTCG); 16SII Fw (TAATACGACTCACTATAG GGGAGTACGGCCGCA) and 16SII Rv (CCCCCGGTGAGGTTCC); 16SIII Fw (TAATACGACTCACTATAGGGAGTACCCGCGCGTCATC) and 16S-3′Rv; 16S-5′Fw and 16SI Rv; 16SI Fw and 16SII Rv; 16SII Fw and 16S-3′Rv, respectively. The relevance of the PCR products was confirmed by sequencing. A linearized pUC18-based plasmid carried a recombinant sR47 gene (Nolivos et al. 2005). These DNA templates served for RNA production by in vitro transcription using a TranscriptAid T7 High Yield Transcription kit (Thermo Fisher Scientific); RNAs were column- (Zymo Research) or PAGE-purified.
In vitro methylation assay
One micromolar of P. abyssi aFib–Nop5 proteins together with 1 μM test RNA were incubated at 72°C (or 37°C where indicated) for 40 min in methylation buffer (50 mM HEPES–NaOH pH 7.9, 150 mM KCl, 10 mM MgCl2) with 100 µM [3H-methyl]-SAM (2 Ci/mmol), obtained by diluting [3H-methyl]-AdoMet (14,1 Ci/mmol, PerkinElmer) with cold AdoMet (Sigma-Aldrich) purified as described in Gerasimaitė et al. (2011). Certain reactions additionally contained varied concentrations and combinations of C/D RNP components, as indicated; the C/D RNP for control assays was assembled by combining 1 µM guide RNA sR47, 3 µM L7Ae, and 2 µM aFib–Nop5 and preincubation at 65°C for 10 min; the cognate substrate tRNA-Leu(CAA) was then added to 1 µM. Aliquots were taken at indicated time points and cooled on ice. Samples were then applied onto Cellulose DE-81 (Whatman) filters; filters were washed with phosphate buffer (50 mM Na-PO4 pH 7.4, 0.01% DEPC) and dried. 3H-methyl incorporation into RNA was determined by scintillation counting in Rotiszint EcoPlus scintillation fluid (Roth). Assays were performed in triplicate at least, and standard deviation is shown as error bars.
HPLC-MS analysis
Methylation reactions for HPLC analysis typically contained 0.5–2 µM test RNA, 1–2 µM aFib–Nop5, and 100 µM SAM, if any. After incubation at 72°C for 40 min, RNA was column-purified (Zymo Research). For nucleoside analysis, 25 pmol test RNA was digested by incubation with nuclease P1 (0.5 u, Sigma-Aldrich) and SAP phosphatase (0.5 u, Fermentas) in P1 buffer (10 mM NaOAc pH 5.2, 1 mM ZnOAc) for 3 h at 37°C. After enzyme inactivation at 75°C for 10 min, RNA hydrolyzate (10 pmol) was loaded onto an integrated HPLC/ESI-MS Agilent 1200 series system equipped with a Discovery C18 column (752.1 mm, Supelco) and eluted with a linear gradient of solvents A (20 mM ammonium formate, pH 3.5) and B (80% aqueous methanol) at a flow rate of 0.3 mL/min at 30°C as follows: 0–20 min, 0%–20% B; 20–22 min, 20%–100% B. For RNase A or T1 digestion analysis, 70 pmol test RNA was incubated with RNase A (20 µg, Thermo Fisher Scientific) in 10 mM Tris-HCl pH < 7, 20 µM EDTA, 10 mM NaCl for 1 h at 60°C; or RNase T1 (3400 u, Thermo Fisher Scientific) in 50 mM Tris-HCl pH 6, 1 mM EDTA for 1 h at 57°C. Both reactions were further supplemented with phosphatase SAP (2 u, Fermentas) and incubated for 2 h at 37°C. Of note, 60 pmol of digested RNA was loaded onto an integrated HPLC/ESI-MS Agilent 1200 series system equipped with a Zorbax C18 column (50 × 2.1 mm, Supelco) and eluted with a gradient of solvents A (5 mM ammonium acetate, pH 7.4) and B (5 mM ammonium acetate pH 7.4, 99% methanol) as follows: 0–15 min, 0%–15% B; 15–20 min, 15%–25% B; 20–20.5 min, 25%–100% B. High-resolution mass spectra of the HPLC-separated RNA digestion products were acquired on a Q-TOF 6250 mass spectrometer (Agilent) equipped with a Dual-ESI source. P1 digestion products were analyzed on a positive ionization mass spectrum, whereas the products of RNases A and T1 were analyzed on a negative.
For further localization of methylated nucleotides, RNase H digestion was used. After the methylation reaction, the mixtures containing 50 pmol of either 16S.5′ or 16S.C were supplemented with 0.3 mM EDTA and 200 pmol of deoxyoligonucleotides (Metabion) that hybridize in between the regions of predicted methylation sites: 16S.5′.1 Rv GACTCGCATGGCTTAGTCGG and 16S.5′.10 Rv CGGCTGCCACCGGCCTTGCC for 16S.5′, and 16C.3′ Rv TTAACGGCTTCCCTACGG for 16S.C. The primers were annealed by heating the mixtures at 95°C for 3 min and slowly cooling to 50°C. Further, 2.5 u of RNase H were added and incubated for 1 h at 50°C. The resulting RNA fragments were column- (Zymo Research) and PAGE-purified. Further digestion with P1 nuclease and HPLC-MS analysis was performed as described above.
Reverse transcription analysis
Reverse transcription analysis was performed following described procedures (Motorin et al. 2007). Of note, 1–2 μM 16S.5′ or full-length 16S rRNA was incubated with 1–2 μM aFib–Nop5 and 100 μM SAM at 72°C for 40 min. For mapping Gm47, 16S.5′ was then processed with RNase H, the 1–93th position fragment was gel-purified and further used for the analysis. Cm847 was mapped on a full-length 16S rRNA that had been column-purified (Zymo Research) after incubation with aFib–Nop5. For alkaline hydrolysis assay, 5 pmol of RNA was incubated with 50 mM Na2CO3 at 90°C for 30 min in a total volume of 10 µL, and then ethanol-precipitated. Of note, 2 pmol 33P-5′-end-labeled primer, 16S.5′.1 Rv for Gm47 and RTc (GGCCGTACTCCCCAGG) (Metabion) for Cm847 mapping, was hybridized to 1 pmol test RNA. For analysis of alkaline hydrolysis, 1 pmol primer and 5 pmol RNA were used; for sequencing reactions 3.5 pmol primer was hybridized to 1.75 pmol RNA (unmethylated). An extension step was performed by addition of 0.01–4 μM dNTP (or 1 mM after alkaline hydrolysis; or 0.3 of the total reaction volume of Termination Mix G/A/T/C [Fermentas] for sequencing), 1/15 of the total reaction volume of RevertAid Premium Enzyme Mix (Fermentas) and incubation at 50°C for 30 min, followed by 5 min at 85°C. After treatment with RNase H, DNA was resolved on a denaturing PAA gel and visualized by radioautography.
RNA structure probing with dimethyl sulfate (DMS) was adapted from Tijerina et al. (2007). Five picomoles of 16S.5′, 16S.C, or 16S were preincubated at 72°C for 5 min in methylation buffer and then treated with 0.5% (of the final volume) DMS for 5 min at 72°C. The reaction was quenched with 20 volumes of 30% β-mercaptoethanol in 0.3 M sodium acetate, and the RNA was ethanol-precipitated. Reverse transcription was performed with 1 pmol of primers (Metabion) Rvf1 GACGCCCCCTTGACTCGCAT on 16S.5′, Rvf2 CTACGGCCGCTTTAGGCCCA on 16S, and Rvf3 TTTCAGCCTTGCGGCCGTAC on 16S.C as described above.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Audronė Rukšėnaitė for her help with HPLC-MS analysis; to Didier Flament for providing P. abyssi cell paste; to Justas Dapkūnas and Jonas Švedas for their help with the computational analysis and to Jaunius Urbonavičius for fruitful discussions. This work was supported by grants from the European Social Fund under the Global Grant measure (VP1-3.1-SMM-07-K-01-105), the Lithuanian State Science and Studies Foundation, the French Ministry of Foreign and European affairs under the Lithuania–France bilateral collaboration programme Gilibert (SUT-157), and the Centre National de la Recherche Scientifique with additional funding from the Agence Nationale de la Recherche (BLAN08-18329396).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059832.116.
REFERENCES
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol 10: 256–263. [DOI] [PubMed] [Google Scholar]
- Aittaleb M, Visone T, Fenley MO, Li H. 2004. Structural and thermodynamic evidence for a stabilizing role of Nop5p in S-adenosyl-L-methionine binding to fibrillarin. J Biol Chem 279: 41822–41829. [DOI] [PubMed] [Google Scholar]
- Appel CD, Maxwell ES. 2007. Structural features of the guide:target RNA duplex required for archaeal box C/D sRNA-guided nucleotide 2′-O-methylation. RNA 13: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MIR, Dy DA, Terns RM, Terns MP. 2005. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Gagnon KT, Brown BA II, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ. 2009. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science 325: 1384–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin ML, Bachellerie JP, Clouet-d'Orval B. 2003. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNATrp. Nucleic Acids Res 31: 6524–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower-Phipps KR, Taylor DW, Wang H, Baserga SJ. 2012. The box C/D sRNP dimeric architecture is conserved across domain Archaea. RNA 18: 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J, Nicoloso M, Bachellerie JP. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383: 732–735. [DOI] [PubMed] [Google Scholar]
- Clouet d'Orval B, Bortolin ML, Gaspin C, Bachellerie JP. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res 29: 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Omer A, Lowe T. 2001. A guided tour: small RNA function in Archaea. Mol Microbiol 40: 509–519. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Tripp V, Lui L, Lowe T, Randau L. 2015. C/D box sRNA-guided 2′-O-methylation patterns of archaeal rRNA molecules. BMC Genomics 16: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Feder M, Pas J, Wyrwicz LS, Bujnicki JM. 2003. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302: 129–138. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Biswas S, Zhang X, Brown BA II, Wollenzien P, Mattos C, Maxwell ES. 2012. Structurally conserved Nop56/58 N-terminal domain facilitates archaeal box C/D ribonucleoprotein-guided methyltransferase activity. J Biol Chem 287: 19418–19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin C, Cavaillé J, Erauso G, Bachellerie JP. 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J Mol Biol 297: 895–906. [DOI] [PubMed] [Google Scholar]
- Gerasimaitė R, Merkienė E, Klimašauskas S. 2011. Direct observation of cytosine flipping and covalent catalysis in a DNA methyltransferase. Nucleic Acids Res 39: 3771–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalei H, Hsiao HH, Urlaub H, Wahl MC, Watkins NJ. 2010. A novel Nop5-sRNA interaction that is required for efficient archaeal box C/D sRNP formation. RNA 16: 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. 2005. Modification and editing of RNA: historical overview and important facts to remember. Top Curr Genet 12: 1–22. [Google Scholar]
- Gulen B, Petrov AS, Okafor CD, Vander Wood D, O'Neill EB, Hud NV, Williams LD. 2016. Ribosomal small subunit domains radiate from a central core. Sci Rep 6: 20885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RH. 1971. The modified nucleosides in nucleic acids. Columbia University Press, New York and London. [Google Scholar]
- Hansen MA, Kirpekar F, Ritterbusch W, Vester B. 2002. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. RNA 8: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Batey RT. 2006. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nucleic Acids Res 34: 5039–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Reyes FE, Batey RT. 2009. Analysis of a critical interaction within the archaeal box C/D small ribonucleoprotein complex. J Biol Chem 284: 15317–15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joardar A, Malliahgari SR, Skariah G, Gupta R. 2011. 2′-O-methylation of the wobble residue of elongator pre-tRNAMet in Haloferax volcanii is guided by a box C/D RNA containing unique features. RNA Biol 8: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Tollervey D. 1998. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci 23: 383–388. [DOI] [PubMed] [Google Scholar]
- Lapinaite A, Simon B, Skjaerven L, Rakwalska-Bange M, Gabel F, Carlomagno T. 2013. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature 502: 519–523. [DOI] [PubMed] [Google Scholar]
- Limbach PA, Crain PF, McCloskey JA. 1994. Summary: the modified nucleosides of RNA. Nucleic Acids Res 22: 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Loza-Muller L, Rodríguez-Corona U, Sobol M, Rodríguez-Zapata LC, Hozak P, Castano E. 2015. Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front Plant Sci 6: 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey JA, Rozenski J. 2005. The Small Subunit rRNA Modification Database. Nucleic Acids Res 33: D135–D138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M. 2011. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2: 611–631. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C. 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53. [DOI] [PubMed] [Google Scholar]
- Muller S, Fourmann JB, Loegler C, Charpentier B, Branlant C. 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:ψ55-synthase and RNA-guided RNA:ψ-synthase activities. Nucleic Acids Res 35: 5610–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Leclerc F, Behm-Ansmant I, Fourmann JB, Charpentier B, Branlant C. 2008. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res 36: 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloso M, Qu LH, Michot B, Bachellerie JP. 1996. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol 260: 178–195. [DOI] [PubMed] [Google Scholar]
- Nolivos S, Carpousis AJ, Clouet-d'Orval B. 2005. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res 33: 6507–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noon KR, Bruenger E, McCloskey JA. 1998. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bacteriol 180: 2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP. 2000. Homologs of small nucleolar RNAs in archaea. Science 288: 517–522. [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Zago M, Chang A, Dennis PP. 2006. Probing the structure and function of an archaeal C/D-box methylation guide sRNA. RNA 12: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H, Robinson H, Terns MP, Terns RM, Yang W, Li H. 2007. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol 371: 1141–1150. [DOI] [PubMed] [Google Scholar]
- Poole A, Penny D, Sjöberg B. 2000. Methyl-RNA: an evolutionary bridge between RNA and DNA? Chem Biol 7: R207–R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakitina DV, Taliansky M, Brown JWS, Kalinina NO. 2011. Two RNA-binding sites in plant fibrillarin provide interactions with various RNA substrates. Nucleic Acids Res 39: 8869–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. 2006. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucleic Acids Res 34: 4293–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Hüttenhofer A. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G, Sommer SS. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8: 404–407. [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DMJ. 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. 2004. Sequential 2′-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 279: 47661–47671. [DOI] [PubMed] [Google Scholar]
- Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T. 2014. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505: 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Mohr S, Russell R. 2007. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc 2: 2608–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkuvienė M, Clouet-d'Orval B, Cerniauskas I, Weinhold E, Klimasauskas S. 2012. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases. Nucleic Acids Res 40: 6765–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Maxwell ES. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J 22: 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Brown J, Maxwell ES. 2004. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem Sci 29: 343–350. [DOI] [PubMed] [Google Scholar]
- Tripp V, Martin R, Orell A, Alkhnbashi OS, Backofen R, Randau L. 2016. Plasticity of archaeal C/D box sRNA biogenesis. Mol Microbiol 103: 151–164. [DOI] [PubMed] [Google Scholar]
- Urbonavičius J, Meškys R, Grosjean H. 2014. Biosynthesis of wyosine derivatives in tRNAPhe of Archaea: role of a remarkable bifunctional tRNAPhe:m1G/imG2 methyltransferase. RNA 20: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65: 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Yang Z, Lin J, Ye K. 2016. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc Natl Acad Sci 113: 10878–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip WSV, Vincent NG, Baserga SJ. 2013. Ribonucleoproteins in archaeal pre-rRNA processing and modification. Archaea 2013: 614735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liang B, Li H. 2011. Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide. RNA 17: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.