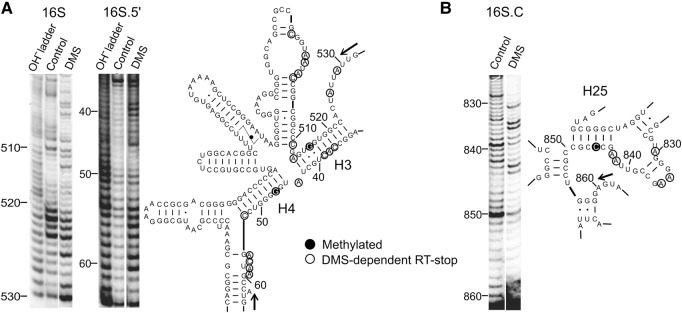
FIGURE 3.
DMS footprinting analysis of RNA secondary structure of aFib–Nop5 target regions of 16S and 16S.5′ RNA (A) and 16S.C (B) (Left panels) probed by DMS. (Right panels) secondary structure models of the full-length P. abyssi 16S rRNA (only areas of interest are shown) (Cannone et al. 2002). Full circles mark aFib–Nop5 methylation targets, open circles mark nucleotides that induce reverse transcription stops after DMS treatment, that is, unpaired cytosines and adenines. Arrows denote the start and direction of the reverse transcription primer extension. Control—no DMS.