Jiang et al. show that ADMETOS interacts with AHL10 and SU(VAR)3–9 homolog SUVH9 and ectopically recruits the heterochromatic mark H3K9me2 to AT-rich transposable elements, causing deregulated expression of neighboring genes.
Keywords: hybrid incompatibility, heterochromatin, imprinted genes, endosperm, transposable elements, polyploidy
Abstract
Hybrid seed lethality as a consequence of interspecies or interploidy hybridizations is a major mechanism of reproductive isolation in plants. This mechanism is manifested in the endosperm, a dosage-sensitive tissue supporting embryo growth. Deregulated expression of imprinted genes such as ADMETOS (ADM) underpin the interploidy hybridization barrier in Arabidopsis thaliana; however, the mechanisms of their action remained unknown. In this study, we show that ADM interacts with the AT hook domain protein AHL10 and the SET domain-containing SU(VAR)3–9 homolog SUVH9 and ectopically recruits the heterochromatic mark H3K9me2 to AT-rich transposable elements (TEs), causing deregulated expression of neighboring genes. Several hybrid incompatibility genes identified in Drosophila encode for dosage-sensitive heterochromatin-interacting proteins, which has led to the suggestion that hybrid incompatibilities evolve as a consequence of interspecies divergence of selfish DNA elements and their regulation. Our data show that imbalance of dosage-sensitive chromatin regulators underpins the barrier to interploidy hybridization in Arabidopsis, suggesting that reproductive isolation as a consequence of epigenetic regulation of TEs is a conserved feature in animals and plants.
Understanding the mechanisms by which populations become reproductively isolated is a major goal of evolutionary biology. Intrinsic post-zygotic isolation results in the sterility or lethality of the F1 hybrid offspring after a successful fertilization event and zygote formation (Presgraves 2010; Maheshwari and Barbash 2011). Post-zygotic reproductive isolation in response to hybridizations of plants that differ in ploidy (interploidy hybridizations) is a major path for sympatric speciation in plants (Ramsey and Schemske 1998; Otto and Whitton 2000). This phenomenon is generally referred to as the “triploid (3×) block,” which describes the difficulty of obtaining viable 3× hybrid plants after diploid (2×)–tetraploid (4×) crosses (Marks 1966). The main determinant for 3× seed failure is abnormal endosperm development (Brink and Cooper 1947; Woodell and Valentine 1961; Ramsey and Schemske 1998; Hehenberger et al. 2012). The endosperm is a nourishing tissue supporting embryo growth, similar to the placenta in mammals. In most flowering plant species, the endosperm is a 3× tissue that develops after fertilization of the homodiploid central cell by one of the two sperm cells, while the second sperm cell fertilizes the egg cell, giving rise to the 2× embryo (Li and Berger 2012). Arabidopsis thaliana follows the most widely distributed mode of endosperm development, where an initial phase of nucleus divisions leads to the formation of a syncytium, which then undergoes cellularization (Costa et al. 2004). Endosperm cellularization is an important developmental transition and essential for embryo survival (Hehenberger et al. 2012). Interploidy hybridizations have an impact on endosperm cellularization, but the effect differs depending on whether increased ploidy is contributed by the maternal or the paternal parent. While hybridizations of maternal plants with pollen donors of increased ploidy delay endosperm cellularization, the reciprocal hybridization causes precocious endosperm cellularization (Scott et al. 1998; Lafon-Placette and Köhler 2016). Interspecies hybridizations cause similar effects on endosperm cellularization (Woodell and Valentine 1961; Ishikawa et al. 2010; Sekine et al. 2013; Rebernig et al. 2015; Lafon-Placette et al. 2017), suggesting that the genes underpinning this hybridization barrier are dosage-sensitive genes (Birchler et al. 2001; Birchler and Veitia 2012). Imprinted genes are expressed predominantly from one of the parental genomes and as a consequence of their monoallelic expression, which is predicted to be dosage-sensitive (Bartolomei and Ferguson-Smith 2011; Patten et al. 2014). It thus has been proposed that imprinted genes are causally involved in interploidy and interspecies seed arrest (Gutierrez-Marcos et al. 2003; Brandvain and Haig 2005; Kinoshita 2007). Consistent with this prediction, loss of function of several paternally expressed imprinted genes (PEGs) restores viability of Arabidopsis 3× seeds by restoring endosperm cellularization (Kradolfer et al. 2013; Wolff et al. 2015).
In this study, we set out to identify the mechanism by which the PEG ADMETOS (ADM) establishes 3× seed arrest (Kradolfer et al. 2013). ADM encodes for a J-domain protein, which is a molecular chaperone involved in diverse biological processes such as protein folding, protein translocation into organelles, and regulation of gene expression by supporting chromatin regulators (Wang and Brock 2003; Dai et al. 2005; Kampinga and Craig 2010). Our data reveal that ADM regulates deposition of histone methylation on Lys9 of histone H3 (H3K9me2), a modification that is connected with heterochromatin formation in plants (Jackson et al. 2002; Malagnac et al. 2002). In 3× seeds, we observed that ADM dependence increased H3K9me2 at small transposable elements (TEs) but reduced levels of CHH (H is any base other than G) methylation, correlating with increased expression of neighboring genes. Our data suggest that mislocalization of H3K9me2 to defined TEs antagonizes the RNA-dependent DNA methylation (RdDM) pathway in 3× seeds, causing deregulated gene expression. Several hybrid incompatibility genes identified in Drosophila encode for heterochromatin-interacting proteins, which has led to the suggestion that hybrid incompatibilities evolve as a consequence of interspecies divergence of selfish DNA elements and their regulation (Johnson 2010; Presgraves 2010; Maheshwari and Barbash 2011). Thus far, evidence supporting this notion has been lacking in plants. Our study shows that the interploidy hybridization barrier is connected to ectopic application of the heterochromatic mark H3K9me2, supporting the view that mechanisms controlling TEs are driving the evolution of hybrid incompatibilities in animals and plants.
Results
ADM interacts with SUVH9 and AHL10
To understand the function of ADM, we identified interacting proteins using a tandem affinity purification (TAP) tagging strategy in an Arabidopsis somatic cell culture (Van Leene et al. 2015). Among the interaction partners that were reproducibly identified in both replicates as interaction partners for ADM were the SU(VAR)3–9 homolog 9 (SUVH9) and seven AT hook-like (AHL) proteins (Supplemental Table S1). SUVH9 binds to methylated DNA and is required for the recruitment of the DNA-dependent RNA polymerase V (Pol V) to chromatin, linking DNA methylation and Pol V transcription in the RdDM pathway (Johnson et al. 2014; Liu et al. 2014). AT hook domain proteins bind to AT-rich chromosomal regions such as matrix attachment regions (MARs) and cause increased levels of H3K9me2 by unknown mechanisms (Yun et al. 2012; Xu et al. 2013). There are two major phylogenetic clades of AT hook proteins in Arabidopsis (Zhao et al. 2014). All AT hook proteins identified as putative ADM interaction partners belong to the B clade (Zhao et al. 2014) and were highly expressed in seeds (Supplemental Fig. S1). Like ADM, AHL10 was up-regulated in 3× seeds (Supplemental Fig. S1; Kradolfer et al. 2013) and was therefore chosen for further analysis.
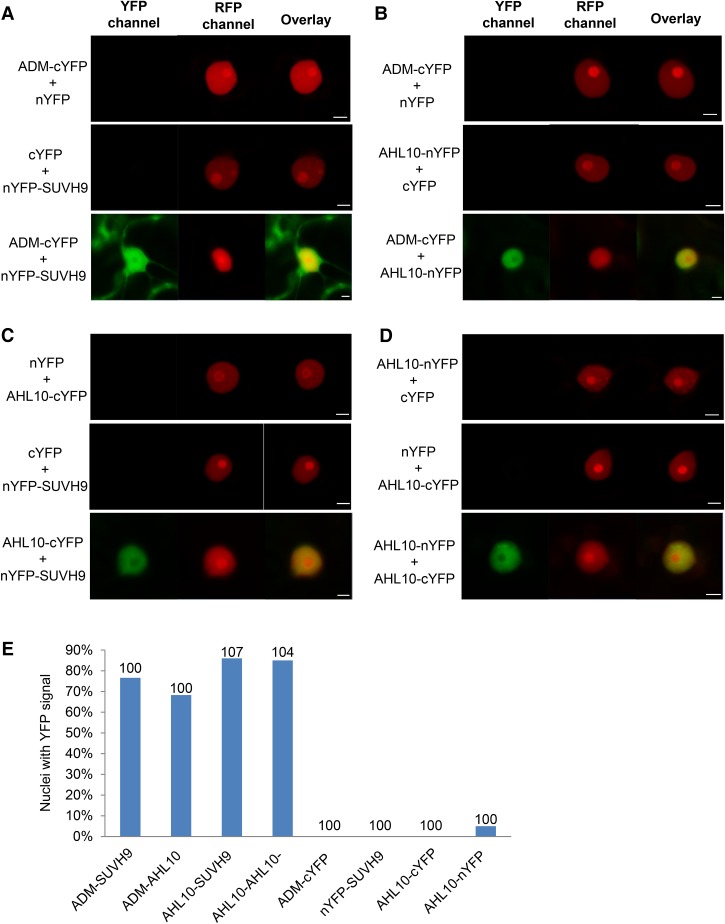
To confirm interaction of ADM, SUVH9, and AHL10, we performed bimolecular fluorescence complementation (BiFC) assays. The results revealed specific interaction of ADM with SUVH9 in the nucleus and cytoplasm (Fig. 1A), consistent with both proteins being localized in both compartments (Supplemental Fig. S2A–G). ADM and SUVH9 interacted with AHL10 in the nucleus (Fig. 1B,C) and AHL10 was also found to form homodimers (Fig. 1D), similar to other AHL proteins (Zhao et al. 2013). We therefore conclude that ADM, AHL10, and SUVH9 may act together to establish the 3× block.
Figure 1.
ADM interacts with SUVH9 and AHL10. Fusion constructs with N-terminal or C-terminal domains of YFP (nYFP or cYFP, respectively) were coexpressed in Nicotiana benthamiana leaves. Vectors expressing nYFP or cYFP alone with each corresponding fusion construct were used as negative controls. H3.1-RFP served as a transformation control. Water-mounted sections of leaf tissue were examined by confocal microscopy. Bars, 5 µm. (A) Interaction of ADM-cYFP with nYFP-SUVH9. (B) Interaction of ADM-cYFP with AHL10-nYFP. (C) Interaction of AHL10-cYFP with nYFP-SUVH9. (D) Interaction of AHL10-nYFP with AHL10-cYFP. (E) The percentage of nuclei showing a YFP signal in the respective combinations of tested interactions. The numbers above the bars correspond to the numbers of scored nuclei.
ADM, SUVH9, and AHL10 establish the 3× block in the endosperm
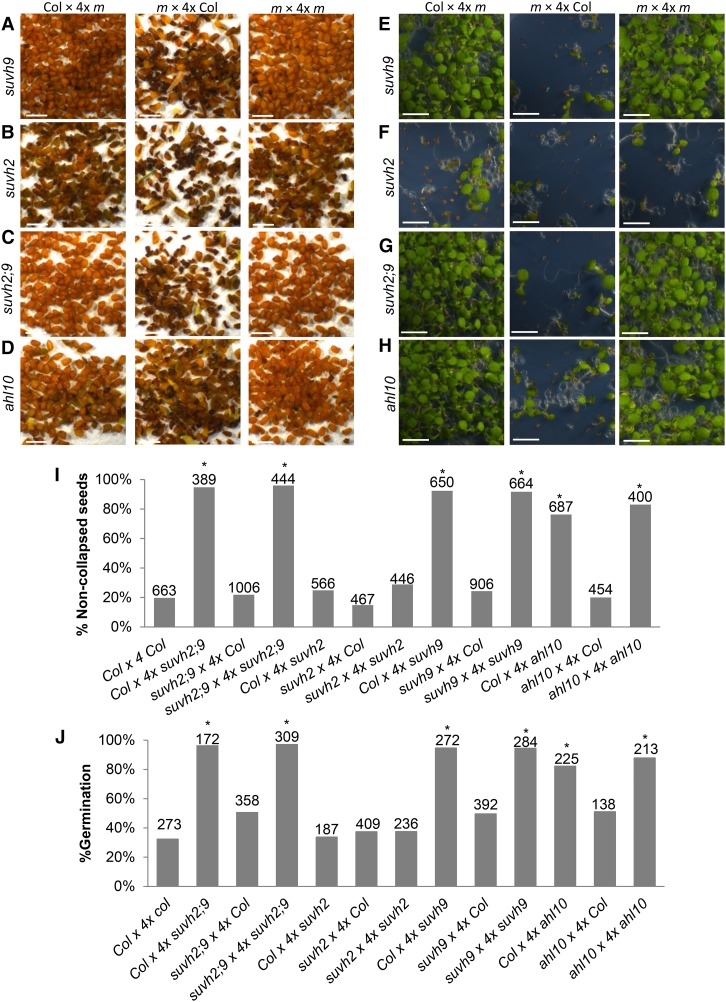
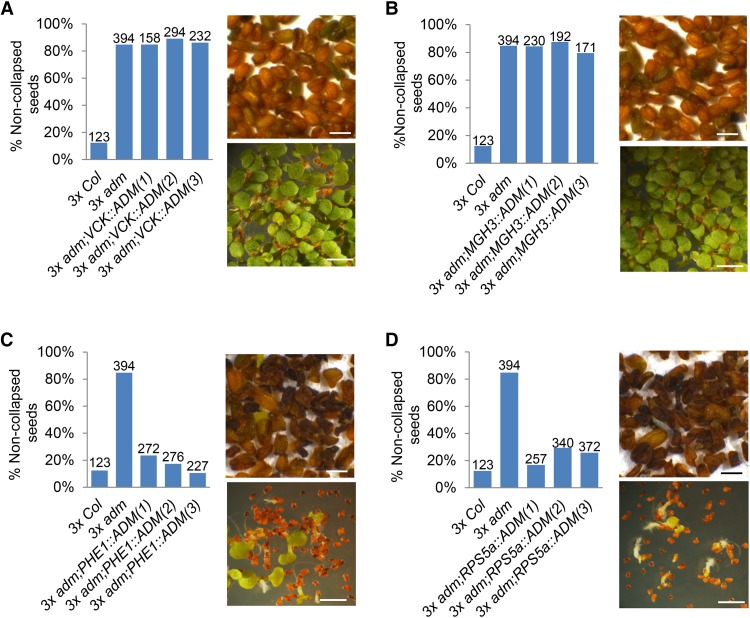
To test the hypothesis that SUVH9 and AHL10 have a role in establishing the 3× block similar to that of ADM, we generated 4× suvh9 and ahl10-1 mutants by colchicine treatment. Since SUVH9 is functionally redundant with SUVH2 in the RdDM pathway (Johnson et al. 2014; Liu et al. 2014), we also generated 4× suvh2 single mutants and suvh2 suvh9 double mutants. Crosses of Col maternal plants with 4× Col pollen donors ( referred to here as Col × 4× Col) gave rise to 20% noncollapsed seeds (Fig. 2A). In contrast, using 4× suvh9, 4× suvh2 suvh9, and 4× ahl10 as pollen donors strongly increased the fraction of noncollapsed and germinating seeds, reaching ∼80% (4× ahl10) to nearly 100% (4× suvh9 and 4× suvh2 suvh9). Pollen of 4× suvh2 did not rescue 3× seed abortion (Fig. 2A–H), consistent with SUVH2 being less expressed in 2× and 3× seeds compared with SUVH9 (Supplemental Fig. S2H). Using suvh9, suvh2 suvh9, and ahl10 as maternal parents did not significantly increase the rate of noncollapsed seeds (Fig. 2A–D,I) and only slightly increased the rate of germinating seeds (Fig. 2E–H,J). Similarly, mutant maternal plants pollinated with mutant pollen resulted in viable 3× seed formation at a frequency similar to the effect when wild-type maternal plants were pollinated with mutant pollen (Fig. 2I,J). We therefore conclude that SUVH9 and AHL10 establish 3× seed abortion when paternally but not when maternally inherited, similar to the paternal effect of ADM (Kradolfer et al. 2013). Like ADM, SUVH9 is a PEG (Pignatta et al. 2014), providing an explanation for the paternal effect of SUVH9. AHL10 had not been identified as an imprinted gene in previous studies (Hsieh et al. 2011; Wolff et al. 2011; Pignatta et al. 2014); however, the expression of AHL10 in the seed coat and low expression at later developmental stages may have confounded its identification. Consistently, we found the maternal AHL10 alleles to be specifically marked by the silencing H3K27me3 modification (Supplemental Fig. S3) that is associated with imprinted expression of PEGs (Moreno-Romero et al. 2016), strongly suggesting that AHL10 is a PEG. In addition, we introduced the suvh9 and ahl10 mutations into the omission of second division1 (osd1) mutant that forms unreduced 2× gametes at nearly 100% frequency (d'Erfurth et al. 2009). Using pollen from either osd1 suvh9 or osd1 ahl10 double mutants strongly suppressed 3× seed abortion, mimicking the effect of 4× suvh9 and ahl10 mutants (Supplemental Fig. S4A–C). We also identified a second mutant allele for AHL10 that, similar to ahl10-1, suppressed 3× seed abortion (Supplemental Fig. S4D). Expression of AHL10 and SUVH9 fusions to the green fluorescent protein (GFP) under control of the UBIQUITIN (UBQ) promoter suppressed the ahl10 and suvh9 mutant phenotype; UBQ::AHL10-GFP nearly completely suppressed viable 3× seed formation, while UBQ::SUVH9-GFP suppressed viable 3× seed abortion to about half the level of the suvh9 mutant (Supplemental Fig. S4E,F). Together, we conclude that SUVH9 and AHL10 are causally responsible for establishing the 3× block in Arabidopsis. The paternal effect of adm, suvh9, and ahl10 on 3× seed rescue made us address the question of whether the proteins act in pollen or the endosperm after fertilization. We therefore expressed ADM under control of a pollen vegetative (VCK) (Grant-Downton et al. 2013) and sperm cell-specific promoter (MGH3) (Okada et al. 2005) as well as the endosperm-specific PHE1 promoter (Weinhofer et al. 2010). While VCK::ADM and MGH3::ADM did not complement the adm mutant, PHE1::ADM conferred complementation to the same level as the RPS5A promoter that is active in embryos and endosperm (Fig. 3A–D; Weijers et al. 2001). Similarly, expression of HA-AHL10 and HA-SUVH9 under control of the PHE1 promoter suppressed formation of viable 3× seeds as efficiently as AHL10-GFP and SUVH9-GFP under control of the constitutive UBQ promoter (Supplemental Fig. S4E,F). These data strongly suggest that ADM, AHL10, and SUVH9 act after fertilization in the endosperm to establish the 3× block.
Figure 2.
3× ahl10 and suvh9 seeds are viable. (A–D) Pictures of mature 3× seeds derived from crosses of the indicated genotypes. (m) Mutant. Bar, 1 mm. (E–H) 3× seedlings derived from crosses of the indicated genotypes at 7 d after germination. Bar, 1 cm. (I) The percentage of noncollapsed 3× seeds derived from crosses of the indicated genotypes. The numbers above the bars correspond to the numbers of analyzed seeds. Asterisks mark significance. χ2 test, P < 10E−10. (J) The percentage of germinated seeds derived from crosses of the indicated genotypes. The numbers above the bars correspond to the numbers of analyzed seeds. Asterisks mark significance. χ2 test, P < 10E−10.
Figure 3.
Rescue of 3× seeds by adm occurs after fertilization. The percentage of noncollapsed 3× seeds derived after pollination with the pollen of osd1, osd1 adm, or osd1 adm expressing ADM under control of the indicated promoters. Three independent transgenic lines were tested for each construct. The numbers above the bars correspond to the numbers of analyzed seeds. At the right of the graph are pictures of mature seeds (top panel) and seedlings at 7 d after germination (bottom panel) of the corresponding genotypes. Bars, 1 cm. (A) osd1 adm mutants expressing the VCK::ADM transgene. (B) osd1 adm mutants expressing the MGH3::ADM transgene. (C) osd1 adm mutants expressing the PHE1::ADM transgene. (D) osd1 adm mutants expressing the RPS5a:ADM transgene.
Interploidy hybridizations cause localization of H3K9me2 to small TEs
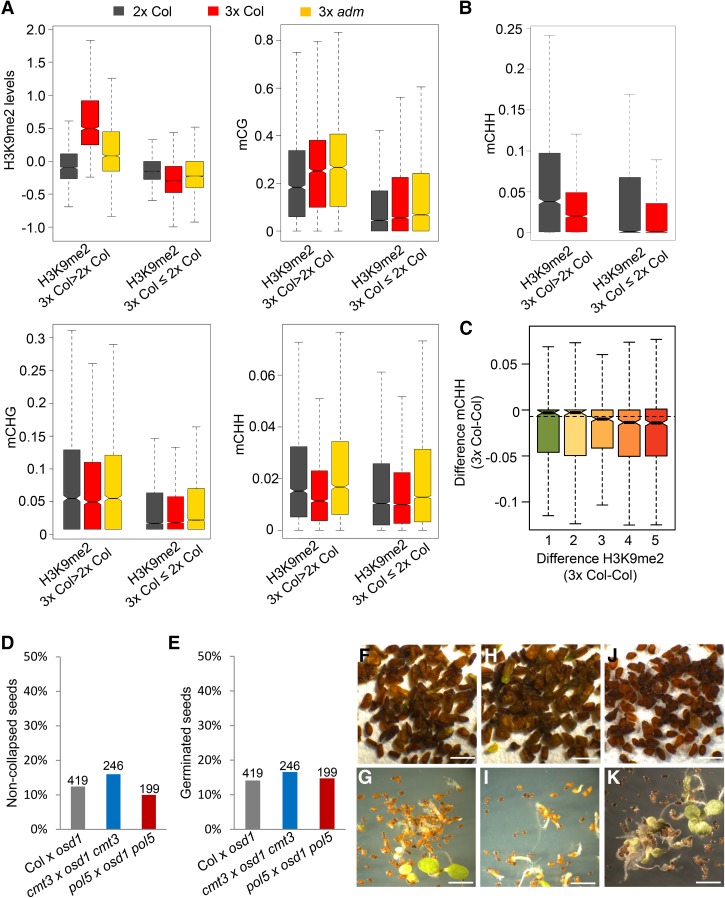
While SUVH9 is not as enzymatically active as H3K9 methyltransferase, it is required for H3K9me2 deposition and likely acts through the RdDM pathway (Johnson et al. 2014; Liu et al. 2014). Also, AHL proteins positively regulate H3K9me2 deposition by unknown mechanisms (Yun et al. 2012; Xu et al. 2013; Zhao et al. 2013). Since expression of ADM, AHL10, and SUVH9 was increased in 3× seeds, we addressed the question of whether this would cause increased H3K9me2 levels in the endosperm of 3× seeds. To test this hypothesis, we generated H3K9me2 profiles of INTACT (isolation of nuclei tagged in specific cell types)-purified endosperm (Moreno-Romero et al. 2016) derived from 2× and 3× wild-type and mutant seeds at 4 d after pollination (DAP) (Supplemental Fig. S5A,B). Previous work proposed that AHL proteins target MARs (Yun et al. 2012; Xu et al. 2013), which are AT-rich DNA elements that specifically bind to the nuclear matrix (Rudd et al. 2004). We therefore tested whether H3K9me2 levels were increased at the 20,000 computationally predicted MARs (Rudd et al. 2004) in the endosperm derived from 3× seeds compared with the endosperm derived from 2× seeds. The level of H3K9me2 was slightly but significantly increased at MARs when comparing the endosperm of 3× versus 2× seeds but also slightly increased at regions not defined as MARs (Supplemental Fig. S6A). The level of H3K9me2 was particularly increased at TEs (Kolmogorov-Smirnov test, P < 10E−15) independently of whether the TEs overlapped with MARs or did not overlap with MARs (Supplemental Fig. S6A). We thus focused further analyses on TEs affected by increased H3K9me2 irrespective of whether they were defined previously as MARs. If ADM is causally responsible for increased H3K9me2 in the endosperm of 3× seeds, H3K9me2 in seeds derived from pollination with osd1 adm pollen should be restored to the level detected in 2× seeds. Indeed, H3K9me2 levels in the endosperm of osd1 adm-derived 3× seeds were substantially reduced compared with osd1-derived 3× seeds (Kolmogorov-Smirnov test, P < 10E−15) (Supplemental Fig. S6A). Likewise, H3K9me2 in the endosperm of 4× suvh2 suvh9-derived and 4× ahl10-derived 3× seeds was substantially reduced compared with 3× wild-type seeds (Supplemental Fig. S6A), supporting the notion that ADM, AHL10, and SUVH9 are causally responsible for increased H3K9me2 levels in the endosperm of 3× seeds. There were no substantial changes in H3K9me2 in 2× seeds derived from pollinations with adm, suvh2 suvh9, or ahl10 pollen (Supplemental Fig. S6B), consistent with adm, suvh2 suvh9, and ahl10 seeds being phenotypically indistinguishable from wild-type seeds (Kradolfer et al. 2013; data not shown).
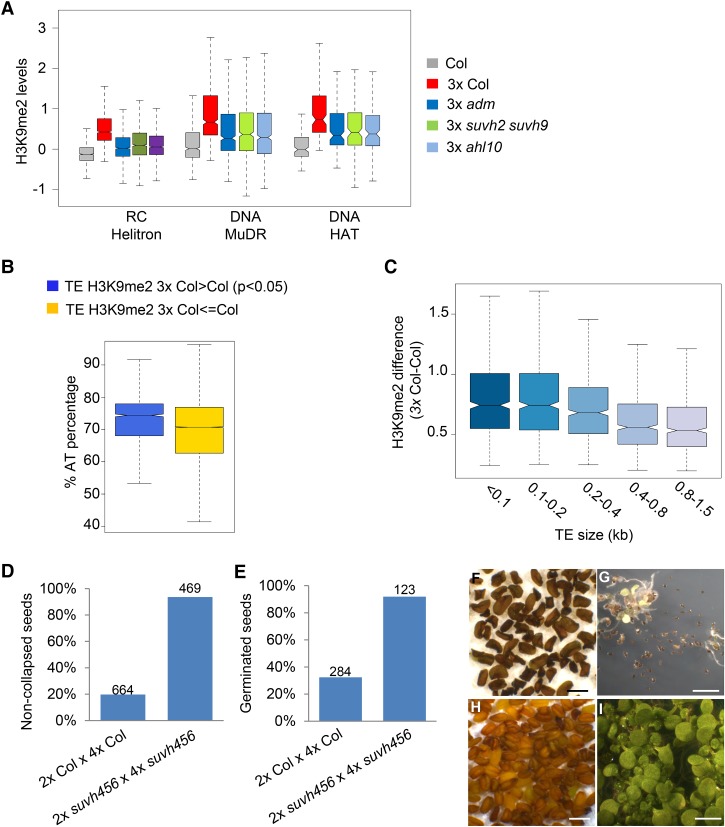
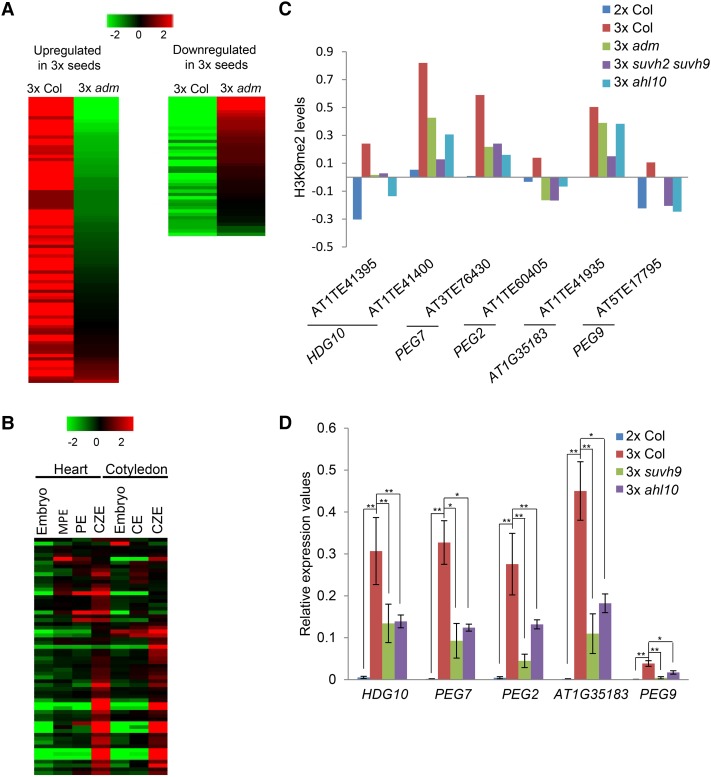
We addressed the question of whether a particular class of TEs is specifically targeted by H3K9me2. We identified 3021 TEs with significantly increased H3K9me2 levels in 3× compared with 2× seeds (P < 0.05). The median size of these TEs was 303 base pairs (bp), with more than half (65%) being >500 bp in size. Nearly all of those TEs had decreased H3K9me2 levels in 3× adm (94%), suvh2 suvh9 (88%), and ahl10 (93%) seeds (Supplemental Table S2). Among them, small AT-rich TEs belonging to RC/helitron, DNA MuDR, and DNA/HAT families were significantly enriched (χ2 test, P < 0.05) (Fig. 4A–C), consistent with the proposed function of AHL proteins to target AT-rich regions (Yun et al. 2012; Xu et al. 2013). Together, we conclude that ADM, AHL10, and SUVH9 promote H3K9me2 deposition at small AT-rich TEs in response to interploidy hybridization in Arabidopsis.
Figure 4.
ADM-dependent ectopic application of H3K9me2 in 3× seeds. (A) Box plots showing median values of z-score-normalized H3K9me2 at TEs belonging to the indicated families in the endosperm of seeds derived from crosses Col × Col (Col), Col × osd1 (3× Col), Col × osd1 adm (3× adm), Col × 4× suvh2 suvh9 (3× suvh2 suvh9), and Col × 4× ahl10 (3× ahl10). Shown are TEs that significantly gain H3K9me2 in 3× Col. limma, P < 0.05. Reduced H3K9me2 in the endosperm of the indicated mutants compared with 3× Col is significant. Kolmogorov-Smirnov test, P < 10E−5. (B) Box plots showing median values of AT percentage within specified groups of TEs. The difference is significant. Kolmogorov-Smirnov test, P < 10E−15. (C) Box plots showing median values of H3K9me2 differences within TEs of the indicated sizes. The difference between the first and last two categories is significant. Kolmogorov-Smirnov test, P < 10E−15. (A–C) Boxes show medians and the interquartile range, and error bars show the full range excluding outliers. (D) The percentage of noncollapsed seeds of the indicated genotypes. The numbers above the bars correspond to the numbers of analyzed seeds. (E) The percentage of germinated seeds of the indicated genotypes. The numbers above the bars correspond to the numbers of analyzed seeds. (F) Picture of mature seeds derived from the cross 2× Col × 4× Col. Bar, 1 mm. (G) Picture of 3× Col seedlings 7 d after germination. Bar, 1 cm. (H) Picture of mature seeds derived from the cross 2× Col × 4× suvh456. Bar, 1 mm. (I) Picture of 3× suvh456 seedlings 7 d after germination. Bar, 1 cm.
To challenge the hypothesis that increased H3K9me2 deposition in 3× seeds is causally responsible for 3× seed abortion, we tested whether mutants in the H3K9me2 methyltransferase genes KRYPTONITE (KYP), SUVH5, and SUVH6 are able to suppress 3× seed abortion. The kyp suvh5 suvh6 triple mutant is nearly completely depleted of H3K9me2 (Stroud et al. 2014). 3× seeds lacking KYP, SUVH5, and SUVH6 function completely suppressed 3× seed abortion (Fig. 4D–I), strongly suggesting that increased H3K9me2 deposition negatively impacts 3× seed survival.
Gain of H3K9me2 at TEs coincides with increased expression of neighboring genes in the endosperm
The majority (85%) of TEs gaining H3K9me2 was not expressed in 2× or 3× seeds. Those TEs that were expressed in one of the samples did not substantially differ in their expression in 2× Col, 3× Col, and 3× adm seeds (Supplemental Fig. S7A; Supplemental Table S3), indicating that gain of H3K9me2 did not confer repression. There were 458 genes within 2-kb distance of TEs gaining H3K9me2 in 3× seeds (Supplemental Table S4). Compared with all genes, these 458 genes had increased transcript levels in 3× Col seeds (Kolmogorov-Smirnov test, P < 0.01) (Supplemental Fig. S7B), and 89 genes (19%) had twofold increased transcript levels in 3× seeds compared with 2× seeds (Fig. 5A; Supplemental Table S3), which was significantly more than expected by chance (χ2 test, P = 6.5 × 10−5). In contrast, there was no enrichment for down-regulated genes (9.1%; χ2 test, P = 0.65). TE-neighboring genes with increased expression in 3× Col were more strongly repressed in 3× adm seeds compared with all genes with increased expression in 3× seeds (Kolmogorov-Smirnov test, P < 0.05) (Supplemental Fig. S7C). For more than half of the up-regulated genes (49) their expression was reduced again by at least half in adm 3× seeds (Fig. 5A; Supplemental Table S3), which again was more than expected by chance (χ2 test, P < 0.05). Up-regulated genes and those that became down-regulated in 3× adm seeds were enriched for genes encoding for endomembrane-localized proteins (P < 5E-04 and P < 8E-05, respectively), including four defensin-like proteins, suggesting activation of defense–response pathways in 3× seeds, similar to what was reported to occur in hybrid seeds of A. thaliana and Arabidopsis arenosa (Burkart-Waco et al. 2013). Up-regulated TE-associated genes were preferentially expressed in the chalazal region of the endosperm (Fig. 5B), consistent with this region undergoing substantial overgrowth in 3× seeds (Scott et al. 1998). Together, these data suggest that ectopic H3K9me2 in TEs increased expression of neighboring genes in 3× seeds, while the removal of H3K9me2 in 3× adm seeds reduced the expression of neighboring genes.
Figure 5.
Gain of H3K9me2 at TEs coincides with increased expression of neighboring genes. (A) Heat map of log2 fold expression changes in 3× seeds versus 2× seeds (3× Col) and 3× seeds versus 3× adm seeds (3× adm) of genes flanked by TEs gaining H3K9me2 in 3× seeds. Shown are genes that were either twofold up-regulated in 3× seeds versus 2× seeds (left panel) or twofold down-regulated in 3× adm seeds versus 3× seeds (right panel). 3× seeds and 3× adm seeds were generated using osd1 and osd1 adm as pollen donors crossed to Col maternal plants. (B) Cluster analysis of up-regulated TE-associated genes based on their expression in embryos and the endosperm during heart and cotyledon stages of seed development. Each row represents a gene, and each column represents a tissue type. Tissue types used were embryos, micropylar endosperm (MPE), peripheral endosperm (PE), chalazal endosperm (CZE), and cellular endosperm (CE). Red and green indicate tissues in which a particular gene is highly expressed or repressed, respectively. (C) Z-score-normalized H3K9me2 at TEs flanking the indicated PEGs in the endosperm of Col, 3×Col, 3× adm, 3× suvh2 suvh9, and 3× ahl10 seeds. (D) Quantitative RT–PCR analysis of PEGs in seeds of the indicated genotypes at 6 DAP. Data were normalized to PP2A. Data show results from biological triplicates. (*) P < 0.05; (**) P < 0.01, Student's t-test.
A subset of PEGs has been shown to be causally involved in establishing the 3× block in Arabidopsis (Kradolfer et al. 2013; Wolff et al. 2015; Huang et al. 2017). With the exception of TEs close to PEGs At1G34650 (HDG10) and At1G35183, most TEs in close association with PEGs did not pass the stringent criteria used to identify TEs with increased H3K9me2 (Supplemental Fig. S8). Nevertheless, manual inspection revealed noticeable changes of H3K9me2 levels in TEs close to an additional four PEGs that all had higher levels of H3K9me2 in the endosperm of 3× seeds compared with 2× seeds, which became depleted again in 3× adm seeds (Fig. 5C; Supplemental Fig. S8). Interestingly, among those PEGs affected by changed H3K9me2 levels were PEG2 and PEG9 (Fig. 5C; Supplemental Fig. S8); both genes were shown previously to suppress 3× seed abortion when mutated (Wolff et al. 2015). Consistent with a regulatory role of H3K9me2 in the expression of neighboring genes, the expression of all PEGs was strongly increased in 3× seeds and reduced again in 3× adm seeds (Wolff et al. 2015). We tested expression of these PEGs by quantitative RT–PCR (qRT–PCR) and found their expression similarly reduced in 3× suvh9 and 3× ahl10 seeds (Fig. 5D). Together, our data suggest that the ADM-, AHL10-, and SUVH9-mediated localized increase of H3K9me2 in 3× seeds positively impacts the expression of neighboring genes, including PEG2 and PEG9 that have been causally connected to 3× seed abortion (Wolff et al. 2015).
Gain of H3K9me2 at TEs occurs independently of DNA methylation
SUVH9 links DNA methylation and noncoding RNA transcription, forming a self-reinforcing feedback loop in the RdDM pathway (Johnson et al. 2014; Liu et al. 2014). We therefore tested whether increased H3K9me2 deposition correlates with increased DNA methylation. We generated DNA methylation profiles of INTACT-purified endosperm from seeds derived after pollination with Col, osd1, and adm osd1 pollen at 4 DAP (Supplemental Fig. S5C). We compared the DNA methylation levels of RC/helitron, DNA MuDR, and DNA/HAT TEs gaining H3K9me2 in 3× seeds (2104 TEs out of 3021 TEs gaining H3K9me2) with TEs of the same TE families not gaining H3K9me2. TEs gaining H3K9me2 had significantly higher levels of DNA methylation in all sequence contexts compared with TEs not gaining H3K9me2 (Fig. 6A). Strikingly, CHH methylation levels of those TEs decreased in the endosperm of 3× seeds compared with 2× seeds (Fig. 6B), making it unlikely that increased H3K9me2 levels are a consequence of increased RdDM activity. Importantly, decreased CHH methylation in the endosperm of 3× seeds was restored in 3× adm seeds (Fig. 6A; Supplemental Fig. S9B), suggesting that ADM-mediated H3K9me2 prevents CHH methylation establishment. The activity of the RdDM pathway increases during endosperm development, with low activity detected at 4 DAP and high activity detected at 6 DAP (Jullien et al. 2012; Moreno-Romero et al. 2016). We therefore tested whether ectopic H3K9me2 has an impact on CHH methylation establishment in the 6-DAP endosperm. TEs with increased H3K9me2 in the endosperm of 3× seeds had significantly higher levels of CHH methylation than TEs not gaining H3K9me2. Consistent with the pattern observed in the 4-DAP endosperm, CHH methylation of TEs gaining H3K9me2 became significantly depleted in 3× seeds (Fig. 6B; Supplemental Fig. S9C), suggesting an antagonistic relationship between both marks. In support of this notion, the presence of both marks was anti-correlated; TEs with the highest gain of H3K9me2 experienced the strongest loss of CHH methylation (Kolmogorov-Smirnov test, P < 0.005) (Fig. 6C). Thus, our data indicate that H3K9me2 and CHH methylation have an antagonistic relationship in 3× seeds; TEs gaining H3K9me2 are compromised in CHH methylation establishment. This interpretation is consistent with previous reports revealing inhibition of the RdDM pathway by heterochromatin with its hallmark, H3K9me2 (Schoft et al. 2009; Zemach et al. 2013; Gent et al. 2014). Together, our data suggest that increased H3K9me2 mediated by ADM–AHL10–SUVH9 does not cause increased levels of DNA methylation in the CHG or CHH context but rather inhibits the RdDM pathway.
Figure 6.
Gain of H3K9me2 at TEs in 3× seeds is independent of DNA methylation. (A) Box plots showing medians of H3K9me2 levels and DNA methylation [#C/(#C+#T)] in each sequence context in 4-DAP endosperm derived from seeds of crosses Col × osd1 (3× Col), Col × osd1 adm (3× adm), and Col × Col (2× Col). TEs belonging to RC/helitron, DNA MuDR, and DNA/HAT families that either gained or did not gain H3K9me2 in 3× seeds are shown. The differences in H3K9me2 and CHH methylation between 2× Col and 3× Col and between 3× Col and 3× adm in TEs gaining H3K9me2 in 3× seeds are significant. Kolmogorov-Smirnov test, P < 10E−15. (B) Box plot showing CHH methylation in 6-DAP endosperm derived from 3× (3× Col) and 2× (2× Col) seeds. The difference in TEs gaining H3K9me2 in 3× seeds is significant. Kolmogorov-Smirnov test, P < 10E−15. (C) Box plot showing the difference of CHH methylation between 3× (3× Col) and 2× (2× Col) seeds in relation to increasing H3K9me2 at TEs in 3× seeds (H3K9me2 3× Col > 2× Col). P < 0.05. The boxes show medians and the interquartile range, and the error bars show the full range excluding outliers. The difference between group 1 and group 5 is significant. Kolmogorov-Smirnov test, P < 0.005. (D) The percentage of noncollapsed seeds derived from the indicated crosses. The numbers above the bars correspond to the numbers of analyzed seeds. (E) The percentage of germinated seeds derived from the indicated crosses. The numbers above the bars correspond to the numbers of analyzed seeds. (F,H,J) Pictures of mature seeds derived from the indicated crosses: Col × osd1 (F), cmt3 × osd1 cmt3 (H), and pol5 × osd1 pol5 (J). Bar, 1 mm. (G,I,K) Pictures of seedlings at 7 d after germination: Col × osd1 (G), cmt3 × osd1 cmt3 (I), and pol5 × osd1 pol5 (K). Bar, 1 cm.
If increased H3K9me2 is not connected to increased levels of CHG and CHH methylation, we expected that mutants impaired in establishing non-CG methylation do not rescue 3× seeds. We therefore tested whether a mutant in CHROMOMETHYLASE 3 (CMT3) could rescue 3× seed abortion. CMT3 is a CHG DNA methyltransferase that binds to H3K9me2 and methylates DNA at nearby CHG sites (Johnson et al. 2007; Du et al. 2012). Consistent with the hypothesis, cmt3 did not rescue 3× seed abortion (Fig. 6D–K), revealing that reduction of CHG methylation is not causally connected to 3× seed rescue. Association of Pol V with chromatin requires SUVH2 and SUVH9. Both proteins contain SRA (SET- and RING-ASSOCIATED) domains capable of binding methylated DNA, suggesting that they recruit Pol V through DNA methylation (Johnson et al. 2014; Liu et al. 2014). Consistent with the proposed independency of ADM-mediated increased H3K9me2 and DNA methylation, loss of Pol V function did not rescue 3× seed abortion (Fig. 6D–K). We thus conclude that rescue of 3× seed abortion occurs independently of components linking SUVH2/9 and DNA methylation.
Accession-dependent differences in the 3× block correlate with differences in H3K9me2 deposition
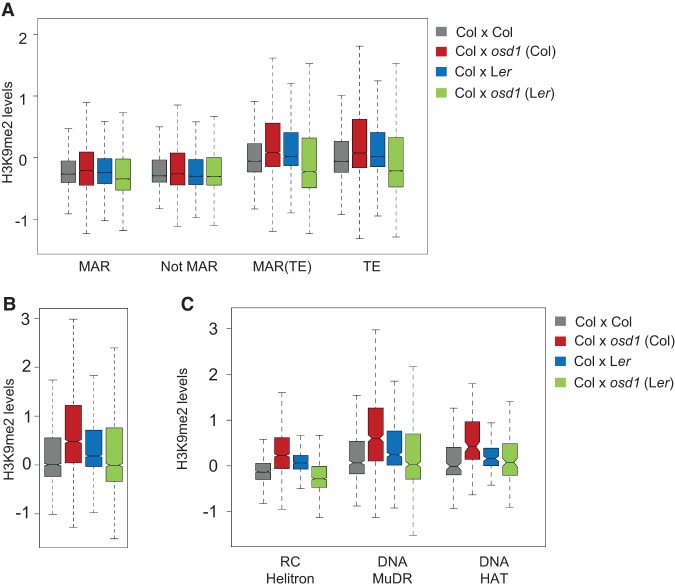
The 3× block is accession-dependent: While the majority of 3× seeds aborts in the Col accession, the majority of 3× seeds is viable in the Ler background (Dilkes et al. 2008). Previously, we found that ADM transcript levels remained much lower in 3× Ler seeds compared with 3× Col seeds (Kradolfer et al. 2013), raising the question of whether reduced ADM transcript levels in 3× Ler seeds correlate with reduced H3K9me2 levels at TEs. Increased viability of 3× seeds derived from 4× Ler pollen donors occurs independently of whether Col or Ler served as the maternal parent (Dilkes et al. 2008). We thus generated H3K9me2 profiles of INTACT-purified endosperm derived from Col × Ler and Col × osd1-2 (Ler) seeds at 4 DAP and asked whether TEs strongly gaining H3K9me2 in 3× Col seeds would have reduced H3K9me2 levels in 3× seeds generated with 2n Ler pollen. We noticed that H3K9me2 levels in the endosperm derived from Col × Ler crosses were higher compared with the endosperm of Col × Col-derived seeds, consistent with previously published data showing higher H3K9me2 levels on nongypsy TEs in Ler compared with Col endosperm (Moreno-Romero et al. 2016). Nevertheless, consistent with ADM not being overexpressed in 3× Ler seeds, H3K9me2 levels did not increase at TEs in Col × osd1-2 (Ler)-derived 3× seeds (Fig. 7A) but rather were reduced compared with 2× seeds. Levels of H3K9me2 were also decreased at those TEs gaining H3K9me2 in 3× Col seeds (Fig. 7B,C), revealing that gain of H3K9me2 at TEs occurs specifically in 3× seeds of the Col but not the Ler accession.
Figure 7.
H3K9me2 is not increased at TEs in 3× Ler seeds. (A) Box plot showing median values of z-score-normalized H3K9me2 in predicted MARs, regions excluding the MAR (Not MAR), predicted MARs overlapping TEs [MAR (TE)], and TEs in the endosperm of the indicated genotypes. All differences are significant. Kolmogorov-Smirnov test, P < 10E−15. (B) Box plot showing median values of z-score-normalized H3K9me2 at TEs significantly gaining H3K9me2 in 3× Col seeds. P < 0.05. Genotypes are specified in A. All differences are significant. Kolmogorov-Smirnov test, P < 10E−15. (C) Box plot showing z-score-normalized H3K9me2 at TEs belonging to the indicated families and significantly gaining H3K9me2 in 3× Col seeds. P < 0.05. Genotypes are specified in A. The difference between 3× Col and 3× Ler is significant. Kolmogorov-Smirnov test, P < 10E−15. The boxes show medians and the interquartile range, and the error bars show the full range excluding outliers.
Discussion
Our study reveals that interploidy hybridizations cause ADM-dependent mislocalization of H3K9me2 to AT-rich small TEs, which is likely causally connected to 3× seed arrest. We found that ADM interacts with AHL10, a member of AT hook domain proteins that have been shown to cause increased levels of H3K9me2 by as yet unknown mechanisms (Yun et al. 2012; Xu et al. 2013). AHL16 (TEK) has been proposed to recruit histone deacetylation complexes (Xu et al. 2013); however, the connection to H3K9me2 remains to be resolved. We identified SUVH9 as an interaction partner for ADM and AHL10, providing a novel link to H3K9me2 establishment by AHL10 and ADM. SUVH9 does not have enzymatic methyltransferase activity but binds to methylated DNA and is required for the recruitment of Pol V to chromatin (Johnson et al. 2014; Liu et al. 2014). Nevertheless, loss of Pol V function did not suppress 3× seed abortion, suggesting that ectopic Pol V recruitment is not a component of the 3× block. Furthermore, CHH methylation did not increase at TEs gaining H3K9me2 but rather decreased, consistent with the notion that SUVH9 is unlikely to recruit Pol V to this subset of TEs. SUVH9 has also been shown to recruit the microrchidia (MORC) adenosine triphosphatase AtMORC6, which is required for transcriptional silencing of TEs and genes independently of the RdDM pathway (Moissiard et al. 2012; Harris et al. 2016). We currently cannot exclude the possibility that AHL10–ADM–SUVH9 ectopically recruit MORC6 or its interacting homologs, MORC1 and MORC2 (Moissiard et al. 2014); nevertheless, all three genes are down-regulated in 3× seeds compared with 2× seeds (Supplemental Fig. S10), which argues against this possibility. Based on our data, we propose that AHL10 recruits ADM and SUVH9, which possibly facilitates recruitment of KYP/SUVH5/SUVH6, leading to increased H3K9me2 at AT-rich TEs. Short AT-rich TEs are generally depleted of H3K9me2 (Zemach et al. 2013), suggesting that the proposed recruitment of KYP/SUVH5/SUVH6 to small AT-rich TEs specifically occurs in 3× seeds as a consequence of increased dosage of AHL10, ADM, and SUVH9.
Consistent with the reported inhibition of the RdDM pathway by heterochromatin (Schoft et al. 2009; Zemach et al. 2013; Gent et al. 2014), we observed a reduction of CHH methylation at TEs gaining H3K9me2. Reduced CHH methylation correlated with increased expression of neighboring genes, in agreement with the observation that DNA-methylated TEs exert a repressive effect on neighboring genes (Hollister and Gaut 2009; Wang et al. 2013). Our data led us to propose that AHL10–ADM–SUVH9-mediated local increase of H3K9me2 inhibits the RdDM pathway, causing increased expression of neighboring genes. H3K9me2 is known to recruit the DNA methyltransferases CMT2 and CMT3 that enforce DNA methylation in a feedback loop with H3K9me2 (Du et al. 2012; Zemach et al. 2013; Stroud et al. 2014). Nevertheless, there are known exceptions to this rule; for example, SUVR5 establishes H3K9me2 independently of DNA methylation (Caro et al. 2012). Recruitment of CMT2 depends on the chromatin remodeler DDM1 that preferentially targets long GC-rich TEs and allows access to chromatin by targeting the linker histone H1 (Zemach et al. 2013). Histone H1 condenses chromatin and inhibits nucleosome mobility and transcription in vitro (Robinson and Rhodes 2006), suggesting that transcriptional inhibition by H3K9me2- and H1-mediated chromatin condensation are functionally connected. The AHL10–ADM–SUVH9-mediated increase of H3K9me2 occurred preferentially at short AT-rich TEs that likely fail to establish histone H1-dependent heterochromatic structures. RdDM depends on the chromatin remodeler DRD1 that facilitates Pol V activity (Law et al. 2010; Zhong et al. 2012). Activity of DRD1 has been suggested to be inhibited by heterochromatic nucleosomes (Zemach et al. 2013), providing a possible explanation of why local increase of H3K9me2 is not connected to increased CHH methylation in 3× seeds.
Our data bear striking similarities to hybrid lethality between Drosophila melanogaster and Drosophila simulans caused by the heterochromatin-interacting proteins hybrid male rescue (Hmr) and lethal hybrid rescue (Lhr). Both proteins form a heterochromatic complex with heterochromatin protein 1a, and, while mutants in both genes are viable, their combination in F1 hybrid males is lethal (Brideau et al. 2006; Satyaki et al. 2014). Hmr and Lhr are not ubiquitous heterochromatin proteins but localize to a specific subfraction of repeat elements (Satyaki et al. 2014), similar to ADM, AHL10, and SUVH9 targeting a specific subset of TEs. Furthermore, Hmr and Lhr are dosage-sensitive (Barbash et al. 2003; Maheshwari and Barbash 2012), mirroring the dosage sensitivity of AHL10, ADM, and SUVH9 identified in this study. TEs coevolve with mechanisms suppressing their activity; therefore, hybridizations of species differing in TE load are expected to cause incompatibilities as a consequence of unleashed TE activity (Blumenstiel 2011; Castillo and Moyle 2012). This idea has been well established in Drosophila by studies revealing that intraspecific crosses result in hybrid dysgenesis as a consequence of increased transcription and transposition of specific TEs in dysgenic germlines (Malone et al. 2015; Kelleher 2016). Similarly, hybridizations of A. thaliana and A. arenosa cause TE activation in the endosperm (Josefsson et al. 2006), suggesting that disruption of the TE silencing mechanism in interspecies hybrids is a conserved feature in animals and plants (Martienssen 2010; Castillo and Moyle 2012). Nevertheless, our data show that differences in dosage or function of chromatin regulators in parental species can also cause off-target effects in hybrids that are not associated with increased TE expression but rather affect neighboring gene expression. A similar effect has been reported in Drosophila hybrids, where Lhr and Hmr cause increased expression of genes located in pericentromeric heterochromatin, while their functional role in the parental species is likely to repress repeat elements (Satyaki et al. 2014).
There are striking similarities between interploidy and interspecies hybrid seed defects, indicative of a common mechanistic basis (Josefsson et al. 2006; Walia et al. 2009; Rebernig et al. 2015; Lafon-Placette et al. 2017). Interploidy hybridizations disrupt the balance of two maternal genomes to one paternal genome in the endosperm, resulting in unbalanced contributions of imprinted genes such as ADM, AHL10, and SUVH9. Our data strongly suggest that ADM, AHL10, and SUVH9 are dosage-sensitive and that their increased dosage in 3× seeds likely causes them to target TEs that are not their natural targets in 2× seeds. A similar effect may occur in interspecies hybrids if the TE load in hybrid seeds does not correspond to the dosage of ADM, AHL10, and SUVH9. Strikingly, chromatin regulators are overrepresented among PEGs identified in plants (Gehring et al. 2011; Waters et al. 2013), and, as demonstrated in this study, several PEGs are part of a common regulatory pathway acting on chromatin. Chromatin regulatory genes are inherently dosage-sensitive (Henikoff 1979; Reuter and Spierer 1992), which possibly makes them prone to become controlled by imprinting mechanisms. While we cannot conclude on the functional role of AHL10, ADM, and SUVH9 in wild-type 2× seeds, it is tempting to propose that they act to re-establish histone modifications after fertilization. Gametic histones are rapidly exchanged after fertilization (Ingouff et al. 2007), implicating a particular requirement of chromatin regulatory proteins to re-establish heterochromatin after fertilization.
In conclusion, our study reveals that interploidy hybridizations cause deregulated expression of imprinted dosage-sensitive chromatin regulators targeting TEs, adding strong support to the hypothesis that genetic conflict between TEs and host fitness drive the evolution of hybrid incompatibility.
Materials and methods
Plant material and growth conditions
Seeds were stratified for 2 d at 4°C and germinated on Murashige and Skoog medium containing 1% sucrose. Plants were transferred to soil after 2 wk and grown in a growth chamber with daily cycles of 16 h of light at 22°C and 8 h of darkness at 18°C. The ahl10-1 mutant T-DNA (SALK_073165) disrupts the ORF at amino acid 126, and the ahl10-2 mutant T-DNA (GABI_141A04) disrupts the ORF at amino acid 28. If not otherwise indicated, ahl10-1 was used for all experiments. The nrpe1-12 (SALK_033852) (Pontier et al. 2005) and cmt3-11 (SALK_148381) (Zhang et al. 2006) mutants were obtained from the Nottingham Arabidopsis Stock Center. The suvh2-2 (Gabi-Kat516A07) (Kuhlmann and Mette 2012), suvh9-1 (SALK_048033) (Johnson et al. 2008), and suvh2 suvh9 (Kuhlmann and Mette 2012) mutants were kindly provided by Markus Kuhlmann. The suvh456 mutant in Col background was obtained from Judith Bender, and osd1-1 introgressed into the Col background and osd1-2 in Ler background (d'Erfurth et al. 2009) from Raphael Mercier. The adm-1 and adm-2 mutants were described previously (Kradolfer et al. 2013). For complementation experiments using different promoters, the adm-1 mutant was used, while the adm-2 mutant was used for ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) and bisulfite sequencing experiments. For crosses, designated female partners were emasculated, and the pistils were hand-pollinated at 2 d after emasculation.
Generation of plasmids and transgenic lines
For the generation of BiFc constructs, ADM, AHL10, and SUVH9 coding sequences were amplified by PCR using primers specified in Supplemental Table S5. The PCR products were cloned into pENTR/D-TOPO (Invitrogen) followed by clonase reaction with pSITE_C_EYFP_N1, pSITE_N_EYFP_N1 (Martin et al. 2009). For the generation of the UBQ10::AHL10-GFP and UBQ10::SUVH9-GFP constructs, AHL10-pENTR and SUVH9-pENTR were cloned into the pUBC-GFP vector (Grefen et al. 2010) by clonase reaction. For the generation of the PHE1::HA-AHL10 and PHE1::HA-SUVH9 constructs, a 3-kb upstream sequence of the PHE1 translational start was cloned into the pB7WG2 vector (Karimi et al. 2002), replacing the 35S promoter. AHL10-pENTR and SUVH9-pENTR were cloned into the pB7WG2 (PHE1 promoter) vector by clonase reaction. The above four constructs were correspondingly introduced into the osd1/+ ahl10/− or osd1/+ suvh9/− double mutant using Agrobacterium tumefaciens-mediated transformation (Clough and Bent 1998). For the generation of MGH3::ADM, VCK::ADM, PHE1::ADM, and RPS5A::ADM constructs, the 900-bp upstream sequence of the VCK translational start, the 1.2-kb upstream sequence of the MGH3 translational start, and the 1.6-kb upstream sequence of the RPS5a translational start were cloned into the pB7WG2 vector (Karimi et al. 2002), replacing the 35S promoter. All constructs were introduced into osd1/+;adm/− mutants, and T1 lines that were homozygous for osd1 were analyzed.
TAP and mass spectrometry
Generation of the C-terminal GSrhino-tagged transgene (Van Leene et al. 2015) and transformation of Arabidopsis cell suspension culture (PSB-L) were carried out as described previously (Van Leene et al. 2011). TAP experiments were performed with 100 mg of total protein extract following published procedures (Van Leene et al. 2011). Protein interactors were identified by mass spectrometry using an LTQ Orbitrap Velos mass spectrometer. Proteins with at least two matched high-confidence peptides were retained. Background proteins were filtered out based on the frequency of the occurrence of the copurified proteins in a large data set containing 543 TAP experiments using 115 different baits (Van Leene et al. 2015).
BiFC assays
Suspensions of A. tumefaciens (strain GV 3101) were infiltrated into leaves of Nicotiana benthamiana as described previously (Martin et al. 2009). At 48 h after infiltration, water-mounted sections of leaf tissue were examined by confocal microscopy (Zeiss, LSM 710). The YFP channel was excited with a 514-nm laser, and the emission signal was collected in a wavelength window between 520 and 582 nm.
RNA extraction and qRT–PCR analysis
Seeds from five siliques were collected for each cross and frozen together with 1.25- to 1.55-mm glass beads (Carl Roth) in liquid nitrogen. The samples were ground in a Silamat S5 (Ivoclar Vivadent) three times for 7 sec. RNA was extracted using the Ambion RNA aqueous phenol-free total RNA isolation kit according to the manufacturer's instructions. Residual DNA was removed using Invitrogen DNase I (Amplification Grade), and cDNA was synthesized using the Fermentas first strand cDNA synthesis kit according to the manufacturer's instructions. qPCR was performed with an iQ5 real-time PCR detection system (Bio-Rad) and Solis BioDyne-5x Hot FIREPol EvaGreen qPCR Mix Plus (ROX, Solis BioDyne) according to the manufacturer's instructions. qPCR was performed with three biological replicates using the primers indicated in Supplemental Table S3, and the results were analyzed as described (Simon 2003) using PP2A as reference gene.
INTACT nucleus purification and ChIP
INTACT nucleus purification and ChIP were performed following previously described procedures (Moreno-Romero et al. 2017). The following antibodies were used in this study: anti-histone H3 (Sigma, H9289) and anti-H3K9me2 (Diagenode, pAb-060-050). All experiments were performed in two biological replicates. Except for cross 2× Col × osd1-2 (Ler), purity of samples was estimated based on spiked-in Ler material as described previously (Moreno-Romero et al. 2017). The osd1-1 mutant was introgressed from Nossen into Col (d'Erfurth et al. 2009) and shared a substantial number of single-nucleotide polymorphisms (SNPs) with Ler except on chromosome 5 (data not shown). We therefore mapped reads of spiked-in Ler material to chromosome 5 to calculate contamination. The amount of spike-in material (Snpc) was calculated as weight of spike-in per weight of sample and corresponded to Snpc = 0.20. The amount of endosperm nuclei in the samples before purification (Enpc) was estimated as Enpc = 0.18. The purity in the interploidy crosses was calculated by adapting the published formula for 2n × 2n crosses to the 4× endosperm of 3× seeds (2:2 maternal:paternal ratio).
DNA extraction and bisulfite conversion
DNA methylation data of the 4-DAP endosperm were generated from INTACT-purified endosperm nuclei of 150–300 mg of 4-DAP INT Col siliques. DNA was purified using the MagJET plant genomic DNA kit (Thermo Fisher Scientific), eluted in 80 µL of elution buffer, and sonicated for 30 cycles (30 sec on and 30 sec off at high power) using a Bioruptor (Diagenode). The bisulfite conversion was performed using the EpiTect Fast DNA bisulfite kit (Qiagen) during the library preparation. Experiments were performed in two biological replicates.
Library preparation and sequencing
Endosperm ChIP-seq libraries were generated using the Microplex library preparation kit (Diagenode) following the manufacturer's protocol with 1 ng of starting material. Bisulfite libraries were generated with the Ovation Ultralow Methyl-Seq library systems (NuGEN) following the manufacturer's protocol using 10 ng of starting DNA. Libraries were sequenced at the SciLife Laboratory (Uppsala, Sweden) on an Illumina HiSeq2000 in 100-bp paired-end fashion. Sequencing reads have been deposited as Fastq files in the Gene Expression Omnibus (GSE100011).
Bioinformatic analysis
Reads of ChIP-seq samples passing a quality control were mapped to the Arabidopsis (TAIR10) genome using Bowtie (Langmead 2010) in single-end mode, allowing for up to two mismatches (Supplemental Table S6). Mapped reads were deduplicated and extended to the estimated average length of the genomic fragments (270 bp). Coverage was estimated and normalized to 10 million reads. H3K9me2 ChIP signals were normalized by subtracting their coverage with H3 ChIP data at every single position in the genome. These data were standardized and normalized for comparative purposes across samples with a z-score (Cheadle et al. 2003) and represented in bedGraph files of 50-bp bins. TEs with differential levels of H3K9me2 between 3× and 2× seeds were identified using linear models as implemented in limma (Ritchie et al. 2015), making use of the information in the two replicates for both conditions. Those TEs with a log2 fold change of >0 and P-value of <0.05 were selected as hypermethylated in 3× seed endosperm.
The 4-DAP bisulfite-treated samples were sequenced in 125-bp paired-end mode. After assessing the quality (Supplemental Table S7) and trimming, each read was split in two 50-bp-long fragments. Splitting the reads and mapping them in single-end mode improved the mapping efficiency substantially in relation to other tested options. Reads were mapped to the TAIR10 reference genome using Bismark (Krueger and Andrews 2011), allowing one mismatch per read. Duplicated reads (aligning to the same genomic position) were eliminated before calculating methylation levels. Cytosine methylation was visualized separately for CG, CHG, and CHH cytosine contexts, with bedGraph files representing average methylation values in 50-bp bins across the genome. The bisulfite conversion rate for each library was calculated based on the methylation status of cytosines in the CHH context from reads mapping to the chloroplast genome.
The data of 6-DAP bisulfite-treated samples have been published previously (Schatlowski et al. 2014). RNA sequencing data used in this study have been published previously (Wolff et al. 2015).
Supplementary Material
Acknowledgments
We thank Cecilia Wärdig for technical support, and Joram van Boven for help with data collection for Figure 1. We thank Markus Kuhlmann for providing seeds of the suvh2 suvh9 double mutant, Raphael Mercier for kindly providing seeds of osd1, and Judith Bender for sharing seeds of the suvh456 triple mutant. Sequencing was performed by the SNP and SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VRRFI), and the Knut and Alice Wallenberg Foundation. This research was supported by a European Research Council Starting Independent Researcher grant (to C.K.), grants from the Swedish Science Foundation (to C.K.), and the Knut and Alice Wallenberg Foundation (to C.K.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.299347.117.
References
- Barbash DA, Siino DF, Tarone AM, Roote J. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP. 2011. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet 27: 23–31. [DOI] [PubMed] [Google Scholar]
- Brandvain Y, Haig D. 2005. Divergent mating systems and parental conflict as a barrier to hybridization in flowering plants. Am Nat 166: 330–338. [DOI] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Brink R, Cooper D. 1947. The endosperm in seed development. Bot Rev 132: 423–541. [Google Scholar]
- Burkart-Waco D, Ngo K, Dilkes B, Josefsson C, Comai L. 2013. Early disruption of maternal-zygotic interaction and activation of defense-like responses in Arabidopsis interspecific crosses. Plant Cell 25: 2037–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro E, Stroud H, Greenberg MV, Bernatavichute YV, Feng S, Groth M, Vashisht AA, Wohlschlegel J, Jacobsen SE. 2012. The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner. PLoS Genet 8: e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM, Moyle LC. 2012. Evolutionary implications of mechanistic models of TE-mediated hybrid incompatibility. Int J Evol Biol 2012: 698198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. 2003. Analysis of microarray data using Z score transformation. J Mol Diagn 5: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costa LM, Gutierrez-Marcos JF, Dickinson HG. 2004. More than a yolk: the short life and complex times of the plant endosperm. Trends Plant Sci 9: 507–514. [DOI] [PubMed] [Google Scholar]
- Dai YS, Xu J, Molkentin JD. 2005. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol Cell Biol 25: 9936–9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. 2009. Turning meiosis into mitosis. PLoS Biol 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilkes BP, Spielman M, Weizbauer R, Watson B, Burkart-Waco D, Scott RJ, Comai L. 2008. The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6: e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. 2012. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S. 2011. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK. 2014. Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Kourmpetli S, Hafidh S, Khatab H, Le Trionnaire G, Dickinson H, Twell D. 2013. Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr Biol 23: R599–R601. [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR. 2010. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Pennington PD, Costa LM, Dickinson HG. 2003. Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos Trans R Soc Lond B Biol Sci 358: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CJ, Husmann D, Liu W, Kasmi FE, Wang H, Papikian A, Pastor WA, Moissiard G, Vashisht AA, Dangl JL, et al. 2016. Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein-coding genes. PLoS Genet 12: e1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C. 2012. Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039. [DOI] [PubMed] [Google Scholar]
- Henikoff S. 1979. Position effects and variegation enhancers in an autosomal region of Drosophila melanogaster. Genetics 93: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res 19: 1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. 2011. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci 108: 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Zhu QH, Zhu A, Wu X, Xie L, Wu X, Helliwell C, Chaudhury A, Jean Finnegan E, Luo M. 2017. Mutants in the imprinted PICKLE RELATED 2 gene, suppress seed abortion of fertilization independent seed class mutants and paternal excess interploidy crosses in Arabidopsis. Plant J 90: 383–395. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. 2007. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037. [DOI] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljic L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. 2010. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol 20: 2137–2143. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T. 2010. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Johnson NA. 2010. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE. 2008. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 4: e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. 2014. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson C, Dilkes B, Comai L. 2006. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16: 1322–1328. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. 2012. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kelleher ES. 2016. Reexamining the P-element invasion of Drosophila melanogaster through the lens of piRNA silencing. Genetics 203: 1513–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T. 2007. Reproductive barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet Syst 82: 177–186. [DOI] [PubMed] [Google Scholar]
- Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C. 2013. An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26: 525–535. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann M, Mette MF. 2012. Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol Biol 79: 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C. 2016. Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol Ecol 25: 2620–2629. [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C, Johannessen IM, Hornslien KS, Ali MF, Bjerkan KN, Bramsiepe J, Glockle BM, Rebernig CA, Brysting AK, Grini PE, et al. 2017. Endosperm-based hybridization barriers explain the pattern of gene flow between Arabidopsis lyrata and Arabidopsis arenosa in Central Europe. Proc Natl Acad Sci 114: E1027–E1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 32: 11.7.1–11.7.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. 2010. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Berger F. 2012. Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195: 290–305. [DOI] [PubMed] [Google Scholar]
- Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. 2014. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet 10: e1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. 2011. The genetics of hybrid incompatibilities. Annu Rev Genet 45: 331–355. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. 2012. Cis-by-trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet 8: e1002597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J. 2002. An Arabidopsis set domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Lehmann R, Teixeira FK. 2015. The cellular basis of hybrid dysgenesis and Stellate regulation in Drosophila. Curr Opin Genet Dev 34: 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks G. 1966. The origin and significance of intraspecific polyploidy: experimental evidence from Solanum chacoense. Evolution 20: 552–557. [DOI] [PubMed] [Google Scholar]
- Martienssen RA. 2010. Heterochromatin, small RNA and post-fertilization dysgenesis in allopolyploid and interploid hybrids of Arabidopsis. New Phytol 186: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162. [DOI] [PubMed] [Google Scholar]
- Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, Stroud H, Husmann D, Zhan Y, Lajoie BR, McCord RP, et al. 2012. MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336: 1448–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Bischof S, Husmann D, Pastor WA, Hale CJ, Yen L, Stroud H, Papikian A, Vashisht AA, Wohlschlegel JA, et al. 2014. Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc Natl Acad Sci 111: 7474–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J, Jiang H, Santos-Gonzalez J, Kohler C. 2016. Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J 35: 1298–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Romero J, Santos-Gonzalez J, Hennig L, Köhler C. 2017. Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat Protoc 12: 238–254. [DOI] [PubMed] [Google Scholar]
- Okada T, Endo M, Singh MB, Bhalla PL. 2005. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J 44: 557–568. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annu Rev Genet 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Patten MM, Ross L, Curley JP, Queller DC, Bonduriansky R, Wolf JB. 2014. The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity 113: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D, Erdmann RM, Scheer E, Picard CL, Bell GW, Gehring M. 2014. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 3: e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez–Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2010. The molecular evolutionary basis of species formation. Nat Rev Genet 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske D. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29: 467–501. [Google Scholar]
- Rebernig CA, Lafon-Placette C, Hatorangan MR, Slotte T, Kohler C. 2015. Non-reciprocal interspecies hybridization barriers in the Capsella genus are established in the endosperm. PLoS Genet 11: e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Spierer P. 1992. Position effect variegation and chromatin proteins. Bioessays 14: 605–612. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D. 2006. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 16: 336–343. [DOI] [PubMed] [Google Scholar]
- Rudd S, Frisch M, Grote K, Meyers BC, Mayer K, Werner T. 2004. Genome-wide in silico mapping of scaffold/matrix attachment regions in Arabidopsis suggests correlation of intragenic scaffold/matrix attachment regions with gene expression. Plant Physiol 135: 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA. 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet 10: e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatlowski N, Wolff P, Santos-Gonzalez J, Schoft V, Siretskiy A, Scott R, Tamaru H, Kohler C. 2014. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell 26: 3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Mosiolek M, Slusarz L, Komnenovic V, Brownfield L, Twell D, Kakutani T, Tamaru H. 2009. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG. 1998. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, Kinoshita T. 2013. Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 76: 792–799. [DOI] [PubMed] [Google Scholar]
- Simon P. 2003. Q-Gene: processing quantitative real-time RT–PCR data. Bioinformatics 19: 1439–1440. [DOI] [PubMed] [Google Scholar]
- Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE. 2014. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Persiau G, Van De SlE, Geerinck J, Van Isterdael G, Witters E, De Jaeger G. 2011. Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol Biol 754: 195–218. [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De SlE, Vercruysse L, Dedecker M, Verkest A, Vandepoele K, et al. 2015. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc 10: 169–187. [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. 2009. Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Brock HW. 2003. Polyhomeotic stably associates with molecular chaperones Hsc4 and Droj2 in Drosophila Kc1 cells. Dev Biol 262: 350–360. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM. 2013. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet 9: e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM. 2013. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci 110: 19639–19644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Geldner N, Offringa R, Jürgens G. 2001. Seed development: early paternal gene activity in Arabidopsis. Nature 414: 709–710. [DOI] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C. 2010. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C. 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Jiang H, Wang G, Santos-Gonzalez J, Kohler C. 2015. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife 4: e10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodell SRJ, Valentine DH. 1961. Studies in British primulas. IX. Seed incompatibility in diploid–autotetraploid crosses. New Phytol 60: 282–294. [Google Scholar]
- Xu Y, Wang Y, Stroud H, Gu X, Sun B, Gan ES, Ng KH, Jacobsen SE, He Y, Ito T. 2013. A matrix protein silences transposons and repeats through interaction with retinoblastoma-associated proteins. Curr Biol 23: 345–350. [DOI] [PubMed] [Google Scholar]
- Yun J, Kim YS, Jung JH, Seo PJ, Park CM. 2012. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J Biol Chem 287: 15307–15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. 2013. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Peng H, Neff MM. 2013. Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci 110: E4688–E4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Qiu J, Roalson EH, Neff MM. 2014. Insights into the evolution and diversification of the AT-hook motif nuclear localized gene family in land plants. BMC Plant Biol 14: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. 2012. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.