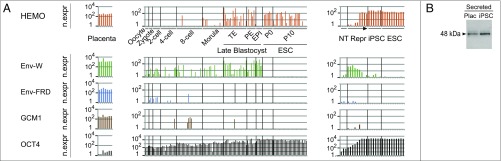
Fig. 7.
Expression of the HEMO gene during development by in silico RNA-Seq analysis. (A) In silico analysis of three panels of RNA-Seq data for HEMO, syncytin-1 (env-W) and -2 (env-FRD), GCM1 (Glial Cells Missing homolog 1, a specific placenta-expressed gene), and OCT4 (highly expressed in stem cells). RNA-Seq raw data were screened with the coding part of each gene, and hits were reported in log scale per kilobase of screened sequence and after normalization with two housekeeping genes, RPLP0 and RPS6 (SI Methods). (Left) Panel of seven samples of normal placental tissues from distinct individuals at the same stage of pregnancy (29). (Center) Panel of 124 single-cell RNA-Seq of human preimplantation embryos and ESCs at the indicated stages of development or cell passage (30) (similar patterns were obtained from data in ref. 31, which covered the oocyte to morula stages). (Right) Panel of 28 RNA-Seq samples from the reprogramming (Repr.; from day 4 to day 21) of human CD34+ cells (NT) to iPSCs (six subclones) and from independent human ESC lines (32). EPI, epiblast; n.expr, normalized expression; P0, Passage 0; P10, Passage 10; PE, primitive endoderm; TE, trophectoderm. (B) Western blot analysis of WGA-purified placental blood (first trimester pregnancy) and WGA-purified supernatant of confluent iPSC cloneN (grown an extra 36 h without serum and concentrated 20×). Samples were treated with PNGase F. The shed HEMO form is detected using the polyclonal anti-HEMO antibody.