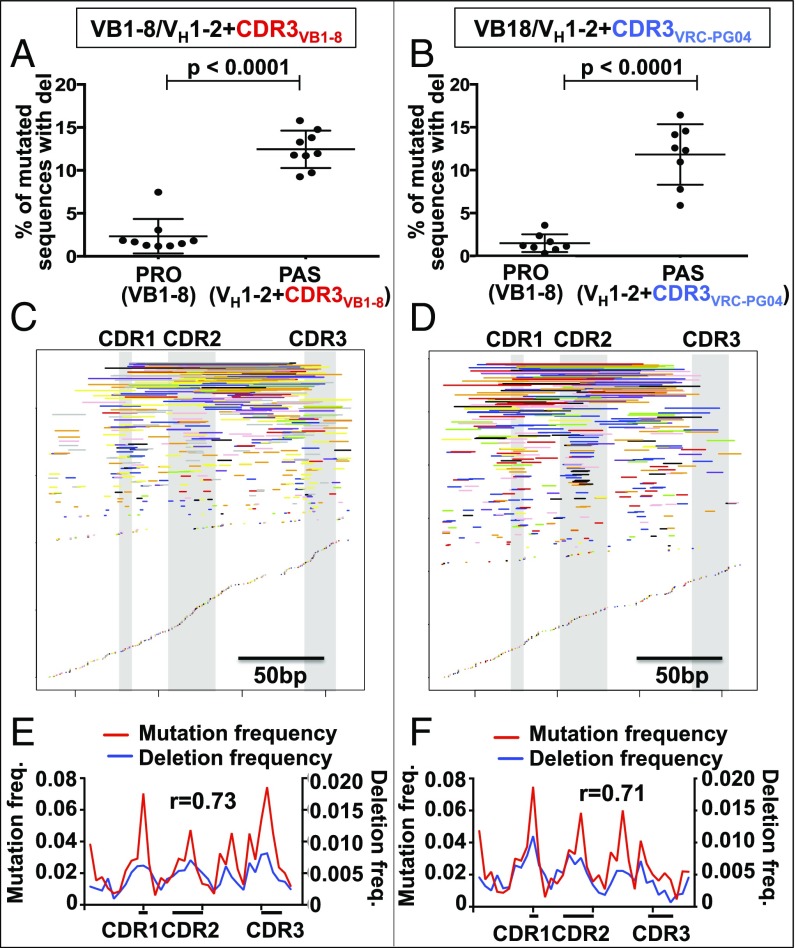
Fig. 2.
Deletions in VH1-2*02 passengers. (A and B) Deletion frequency, calculated as the percent of mutated sequences that contain deletions of (A) VH1-2*02+CDR3VB1-8 and (B) VH1-2+CDR3VRC-PG04 passenger alleles, ± SD (C and D) Map of unique deletions in (C) VH1-2*02+CDR3VB1-8 and (D) VH1-2*02+CDR3VRC-PG04 passenger allele sequences. Deletions are represented by lines whose start and end indicate the start and end of the deletion. Deletions from each of the 9 VH1-2*02+CDR3VB1-8 and 8 VH1-2*02+CDR3VRC-PG04 chimeras are displayed with a different color. (E and F) The location of SHMs compared with the location of deletion endpoints. r is the Pearson correlation coefficient between SHM frequency and deletion endpoint frequency of each bin. n = 9 VB1-8/VH1-2*02+CDR3VB1-8 and 8 VB1-8/VH1-2*02+CDR3VRC-PG04 chimeras.