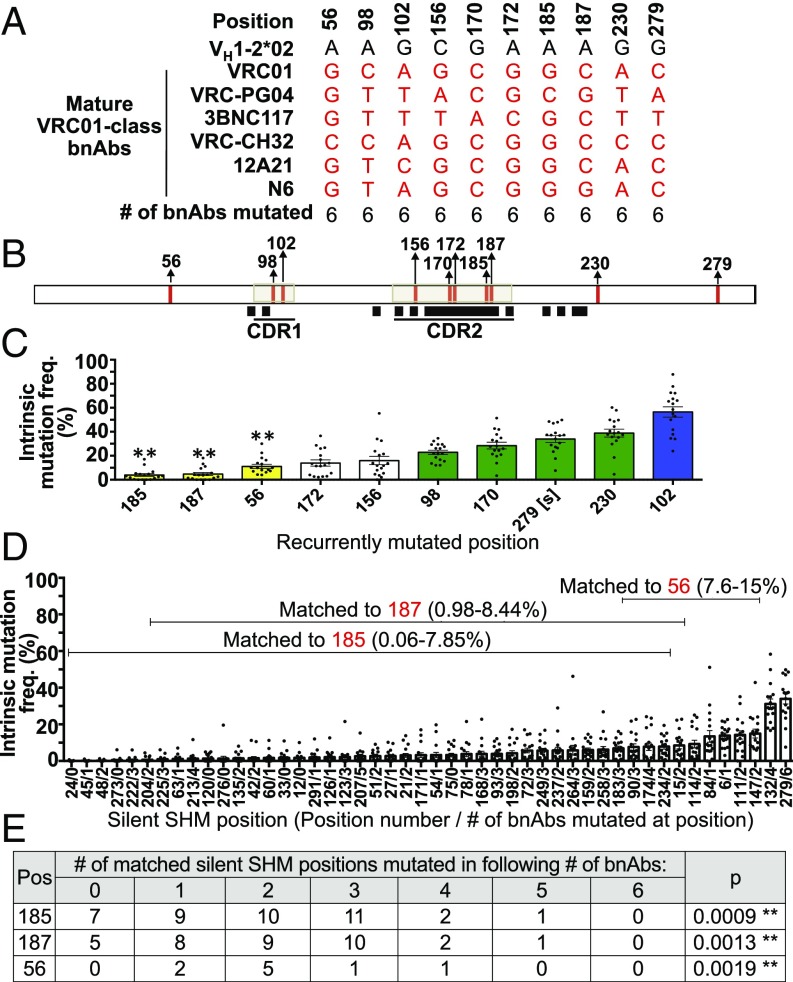
Fig. 3.
Recurrently mutated VH1-2*02 nucleotides among VRC01-class bnAbs. (A) Recurrently mutated VH1-2*02 positions among the IgH V region exon sequences of potent VRC01-class bnAbs VRC01 (14), VRC-PG04 (14), 3BNC117 (14), VRC-CH32 (21, 26), 12A21 (14), and N6 (31). (B) Locations within the VH1-2*02 sequence of recurrently mutated bnAb positions (red vertical lines) and of sequences that contribute to VRC01–gp120 contact sites [black squares (16)]. (C) Intrinsic mutation frequency of recurrently mutated bnAb positions calculated from VH1-2*02 passenger sequences of mutational stratum 21–30 mutations per sequence. Data shown as mean ± SEM of nine VB1-8/VH1-2*02+CDR3VB1-8 and eight VB1-8/VH1-2*02+CDR3VRC-PG04 chimeras. [s] indicates that position 279 is a silent SHM position. Blue and green bars indicate primary and secondary hotspots. Yellow bars indicate positions for which there are silent SHM positions of VH1-2*02 of comparable intrinsic mutation frequency. (D) Silent SHM positions of VH1-2*02. y axis indicates intrinsic mutation frequency, calculated as in C. Brackets indicate the set of silent SHM positions that are matched in intrinsic mutation frequency to each of positions 185, 187, and 56. (E) The number of VRC01-class bnAb sequences mutated at silent SHM positions matched in intrinsic mutation frequency to positions 185, 187, and 56. p is the probability of matched silent SHMs being mutated in six of six bnAbs, estimated by modeling the number of silent SHM positions mutated in each number of bnAbs as a binomial distribution. In C and E ** indicates positions with P values < Bonferroni-adjusted α of 0.05/3.