Abstract
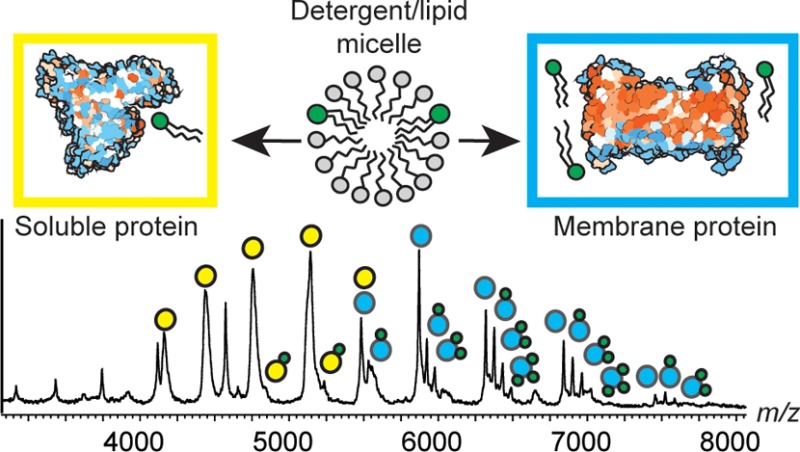
A wide variety of biological processes rely upon interactions between proteins and lipids, ranging from molecular transport to the organization of the cell membrane. It was recently established that electrospray ionization mass spectrometry (ESI-MS) is capable of capturing transient interactions between membrane proteins and their lipid environment, and a detailed understanding of the underlying processes is therefore of high importance. Here, we apply ESI-MS to investigate the factors that govern complex formation in solution and gas phases by comparing nonselective lipid binding with soluble and membrane proteins. We find that exogenously added lipids did not bind to soluble proteins, suggesting that lipids have a low propensity to form electrospray ionization adducts. The presence of detergents at increasing micelle concentrations, on the other hand, resulted in moderate lipid binding to soluble proteins. A direct ESI-MS comparison of lipid binding to the soluble protein serum albumin and to the integral membrane protein NapA shows that soluble proteins acquire fewer lipid adducts. Our results suggest that protein–lipid complexes form via contacts between proteins and mixed lipid/detergent micelles. For soluble proteins, these complexes arise from nonspecific contacts between the protein and detergent/lipid micelles in the electrospray droplet. For membrane proteins, lipids are incorporated into the surrounding micelle in solution, and complex formation occurs independently of the ESI process. We conclude that the lipids in the resulting complexes interact predominantly with sites located in the transmembrane segments, resulting in nativelike complexes that can be interrogated by MS.
Many proteins interact with lipids and membranes in their native environment. Membrane proteins in particular use lipid binding to tune their biological function. These lipid contacts are, however, often transient and heterogeneous, and therefore can be difficult to observe directly. Electrospray ionization mass spectrometry (ESI-MS), which is now widely applied to study noncovalent protein complexes, has recently been adapted to study integral membrane proteins through controlled ejection from detergent micelles in the gas phase.1 By use of suitable detergents in combination with collisional activation, ESI-MS is also able to preserve the interactions between membrane proteins and phospholipids, allowing detailed insights into the interactions with their native lipid environment.1,2 The approach is applicable to lipids that are incorporated into the protein structure as well as annular lipids that modulate protein function or stability.3−5 The range of protein–lipid interactions that can be observed by ESI-MS is surprisingly large: lipids can be copurified with the protein and retained even under denaturing conditions,6−8 but lipids may also be added directly to detergent-solubilized membrane proteins to yield nativelike complexes with defined compositions.9−11 Although only certain lipids are able to exert structural or functional effects, insights from ESI-MS suggest that membrane proteins exhibit a robust ability to bind a large variety of lipids, with the intensity and number of lipid adducts being largely determined by the lipid concentration.9 In this respect, lipid binding to membrane proteins shows hallmarks of nonspecific adduct formation, a common feature in ESI-MS of protein–ligand complexes. Nonspecific electrospray adducts occur when free ligand molecules are concentrated in the same droplet as the protein during the last stages of the electrospray process and are deposited on the protein surface as the solvent evaporates.12−15
Here, we investigate the formation and preservation of protein–lipid complexes in ESI-MS by comparing the lipid-binding abilities of membrane proteins and soluble proteins. We selected three well-understood soluble protein systems: ubiquitin, for which no lipid interactions have been reported; bovine serum albumin (BSA), which has a well-documented propensity to bind lipids nonspecifically in solution in line with its biological role as a carrier protein;16,17 and the N-terminal domain of major ampullate spidroin 1 (MaSp1-NT), which, although highly soluble, shares the α-helical architecture and segregation of charged surface areas common among integral membrane proteins and has been suggested to interact with lipids during storage in the silk gland.18,19 Interestingly, we find that, in all three cases, the lipids do not form nonspecific electrospray adducts and instead require the presence of detergent micelles to associate with proteins. In line with this, direct comparisons show that membrane proteins exhibit a greater lipid-binding propensity than soluble proteins. Our findings suggest that detergent micelles serve as vehicles that mediate the lipid binding, and consequently, the interactions between lipids and membrane proteins evidenced by ESI-MS are reflective of contacts formed in solution rather than simply nonspecific adduct formation.
Experimental Section
Bovine serum albumin (BSA) and ubiquitin were purchased from Sigma. NapA and MaSp1-NT were purified as described.20 Phospholipids (Avanti Polar Lipids, Inc.) were dissolved at approximately 5 mg of dry material in 1 mL of CHCl3 in a glass vial. Lipid films were generated by solvent evaporation under a stream of N2 and dried in a vacuum chamber overnight. Dry lipid films were solvated in deionized H2O by multiple rounds of sonication and vortexing and subsequently lyophilized. The resulting lipid cakes were dissolved in 1 mL of dH2O and centrifuged for 1 h at maximum speed in a benchtop centrifuge. The supernatant was subjected to phosphate analysis to determine the lipid content, revealing a final lipid concentration between 100 and 500 μM, and was stored in 50 μL aliquots at −20 °C.
Prior to MS analysis, ubiquitin, BSA and MaSp1-NT were desalted by exchange into 1 M ammonium acetate, pH 7.5, using BioSpin microcentrifuge columns. For soluble proteins, detergents were added after buffer exchange at the concentrations indicated below. Critical micelle concentration (cmc) values were taken from le Maire et al.21 NapA samples were subjected to solvent and detergent exchanges into 100 mM ammonium acetate pH 7.5 containing 0.5% tetraethylene glycol monooctyl ether (C8E4) at 4 °C in a Superdex Increase 200 column on an Äkta Purifier FPLC system (GE Healthcare). MS analyses were conducted at a protein concentration of approximately 10 μM and a lipid concentration of approximately 50 μM unless noted otherwise.
Samples were introduced into the mass spectrometer by use of gold-coated borosilicate capillaries produced in-house. Mass spectra were recorded on a Waters Synapt G1 mass spectrometer (Waters, Milford, MA) adapted by the manufacturer for high mass analysis and operated in time-of-flight (ToF) mode unless noted otherwise. Instrument settings were as follows: capillary voltage 1.5 kV, sample cone voltage 30 V, extraction voltage 4 V, collision voltages in the trap ranging between 10 and 130 V, bias voltage 5 V, and transfer collision voltage 10 V. The source pressure was 5 mbar. Trap gas was N2 with a flow rate of 6 mL/h for BSA and NapA and 2 mL/h for ubiquitin and MaSp1-NT. For ion mobility (IM) mass spectrometry of BSA, T-wave velocity was 350 m/s and wave height was 10 V in the IMS cell. The transfer wave velocity was 248 m/s and the transfer wave height 13 V. IMS cell gas was N2 with a pressure of 1.6 Torr. Data analysis was performed with the Waters MassLynx 4.1 and PULSAR software packages.22
Results
As a first step, we optimized the ESI-MS conditions for the soluble proteins ubiquitin, serum albumin, and MaSp1-NT for maximal preservation of noncovalent ligand interactions. This was done by raising the source pressure to 5 mbar, reducing the cone voltage to 30 V, and setting the trap and transfer voltages to 10 V.23 Under these conditions, we observed narrow charge-state distributions for all proteins, as well as moderate peak broadening attributable to salt adducts that could be removed by raising the collision voltage in the trap region to 50 V.
Having established suitable MS conditions, we attempted to observe protein–lipid interactions using a direct ESI-MS binding assay.24 For this purpose, the anionic phospholipid 1-palmitoyl-2-oleoyl-glycero-3-phosphoglycerol (POPG) or the zwitterionic phospholipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) or 1-hexadecanoyl-2-(9Z-octadecenoyl)-glycero-3-phosphoethanolamine (POPE) were prepared according to the standard solubilization protocol (see Experimental Section)25 and added directly to the protein in buffered ammonium acetate solution. In all cases, lipids were present in a final concentration of 20-fold excess over the protein. First, we tested ubiquitin, for which no lipid interactions are reported. No peaks were observed corresponding to the mass of the protein with one or more lipid adducts. We then applied the same approach to serum albumin, which is a lipid carrier, and MaSp1-NT, which has charged as well as hydrophobic surfaces, but we did not detect any lipid binding in either case (Figure 1, red traces). Lipids were added to the integral membrane protein NapA in the detergent tetraethylene glycol monooctyl ether (C8E4), a poly(ethylene glycol) (PEG) detergent with a mildly polar head composed of four ether groups. C8E4 can easily be removed at low activation energies and in this manner facilitates MS analysis under gentle ionization conditions, while “milder” detergents like N-dodecyl β-d-maltoside (DDM) remain bound at high activation energies (Figure S1).26 When analyzed under identical MS conditions, all three phospholipids (POPG, POPE, and POPC) were found to bind to the protein (Figure S2). We then added C8E4 to the soluble proteins to match the solvent conditions used for membrane proteins. At 2× CMC, the detergent caused moderate charge reduction but had no adverse effects on peak intensity or the width of the charge-state distributions of ubiquitin, serum albumin, or MaSp1-NT. We then repeated the direct binding assay by adding POPG directly to the ubiquitin solution containing 2× cmc C8E4 (Figure 1, green traces). Surprisingly, the spectra showed an adduct peak corresponding in mass to the protein with a POPG molecule bound. Similarly, peaks indicating binding of one POPG lipid could be observed for serum albumin in the presence of C8E4. Lipid addition to MaSp1-NT with C8E4 resulted in the appearance of peaks indicative of one, two, or three bound POPG molecules. Repeating the experiment with the phospholipids POPE and POPC, we found that only minor amounts of POPC and no POPE remained bound to MaSp1-NT in the gas phase. Ubiquitin exhibited a similar pattern, readily retaining POPG and minor amounts of POPC, while serum albumin bound only to POPG. By contrast the integral membrane protein NapA was found to bind all three lipids, although POPG addition again resulted in more intense adduct peaks than the addition of POPC or POPE (Figure S2).
Figure 1.
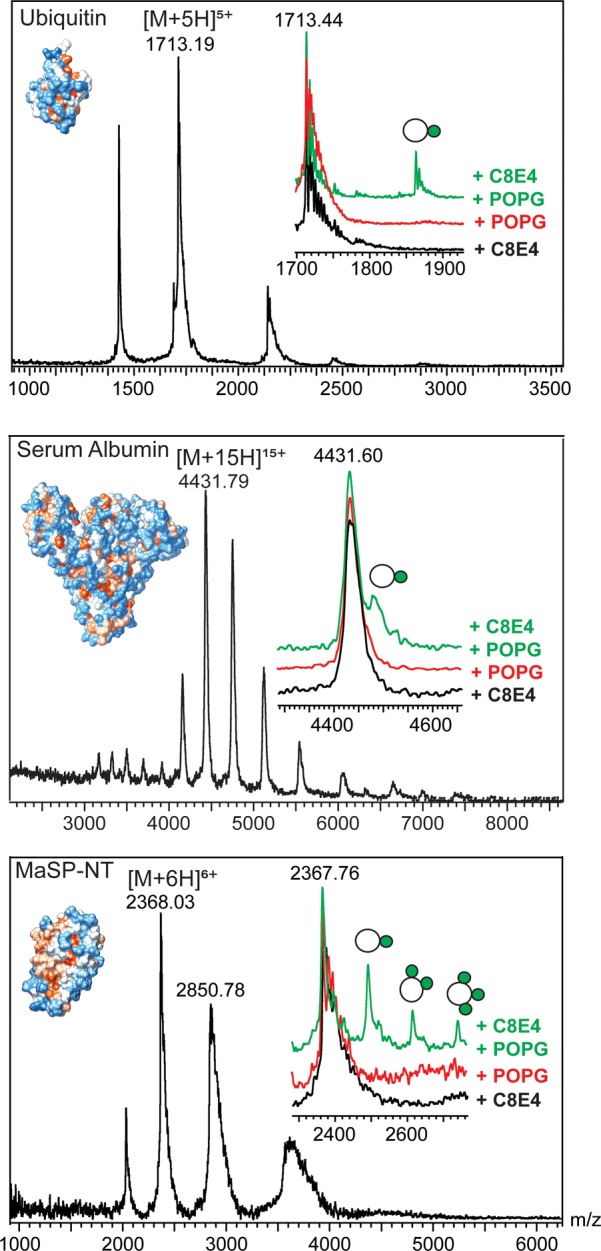
Detergent micelles promote the formation and detection of protein–lipid complexes in ESI-MS. Phospholipid binding to (top) ubiquitin, (middle) BSA, and (bottom) MaSp1-NT can be observed in the presence of detergent C8E4. (Insets) Respective main charge state, with lipid adducts indicated. Protein structures are colored according to hydrophobicity, with blue denoting hydrophilic and orange denoting hydrophobic surface areas. PDB IDs are listed in Table S1.
The crucial role of the detergent in lipid binding led us to speculate that it helps to solubilize the lipids and in this manner increase their accessibility for nonselective association. Examination of the low m/z region revealed large amounts of free lipid monomers and dimers present under even the gentlest MS conditions in the absence of detergent, as well as only a moderate increase in lipid monomers after the addition of detergent (Figure S3A,B). To investigate whether the charge-reducing effects of the detergent promote the detection of protein–lipid complexes, we recorded spectra of MaSp1-NT and POPG in the presence of 2.5% dimethyl sulfoxide (DMSO), which, like C8E4, has been shown to provide charge reduction and preserves noncovalent interactions following ESI-MS.27 Interestingly, addition of DMSO instead of C8E4 did not lead to the formation of protein–lipid complexes (Figure S3C).
Having established that the observed effects are related specifically to the presence of detergent, we selected the interaction between MaSp1-NT and POPG to investigate the effect of detergent concentration while keeping the lipid concentration constant. The presence of submicellar amounts of C8E4 (0.13%) led to the appearance of only minor lipid adducts. Raising the C8E4 concentration to 1× cmc (0.25%) caused a moderate increase in the intensity of the peaks assigned to protein–lipid complexes, while significant lipid binding could only be observed at 2× cmc (0.5%) (Figure 2 A).
Figure 2.
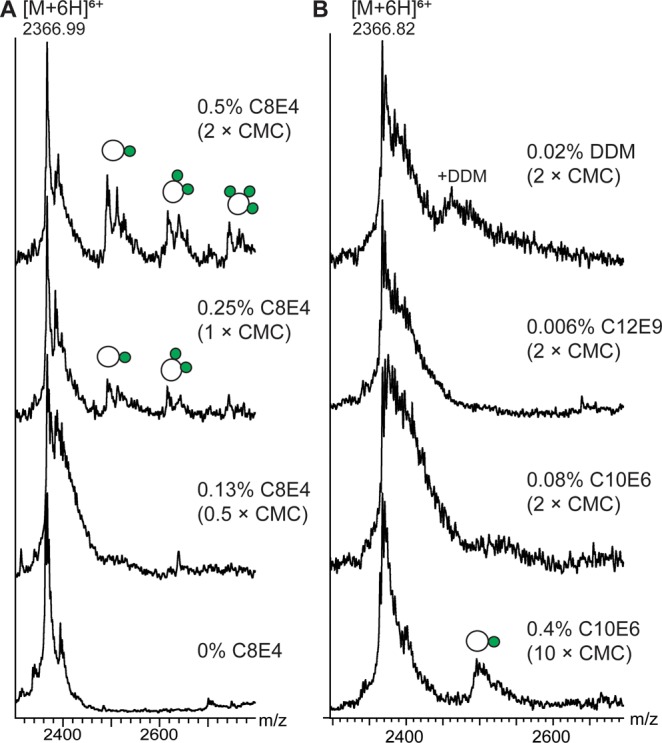
Effective protein–lipid complex formation requires the presence of high amounts of detergent micelles. (A) Complexes between MaSP1-NT and POPG could be observed only in the presence of C8E4 above the cmc. (B) Detergents with lower cmc did not promote binding of POPG to MaSp1-NT. However, when the concentration of C10E6 was increased to that of C8E4, moderate lipid binding could be observed.
We next investigated whether the number of micelles had an effect on lipid binding to MaSp1-NT. Longer chain-length detergents have a greatly reduced CMC, thus forming fewer micelles when these detergents are added at low concentrations. In addition to C8E4, we selected N-dodecyl β-d-maltoside (DDM, cmc 0.009%), C12E9 (cmc 0.003%), and C10E6 (cmc 0.03%). At 2× cmc, the approximate micelle concentrations were 200 μM (C8E4), 27 μM (C10E6), 1 μM (C12E9), and 4 μM (DDM). Interestingly, at these concentrations, none of the three detergents led to the formation of protein–lipid complexes (Figure 2 B). If, however, C10E6 was added at 10× cmc, which effectively increases the micelle concentration to 135 μM, complexes between MaSp1-NT and POPG could be observed. Taken together, this suggests that the formation of protein–lipid complexes is promoted by increasing the number of detergent micelles to the range where nonspecific protein micelle interactions are likely to occur.
Previous studies demonstrated that lipid binding conveys stabilization of membrane proteins against collisional unfolding.9 We therefore investigated the stability of complexes between ubiquitin, serum albumin, or MaSp1-NT and POPG by subjecting them to collisional activation by increasing the voltage in the ion trap of the mass spectrometer. Interestingly, we found that the three protein–lipid complexes exhibited differential stabilities against collisional dissociation: while MaSp1-NT complexes with POPG dissociated readily at low collision voltages (10–50 V), the interaction between serum albumin and a single POPG molecule remained intact well above 100 V (Figure 3 A and Figure S4). We therefore selected this complex to analyze the impact of lipid binding on the conformational stability of the protein by monitoring the change in the arrival-time distribution in response to gas-phase activation.28 We found that the lipid-bound protein was able to retain a compact conformation at higher activation energies, suggesting that POPG binding imparts stabilization against unfolding (Figure 3 B), although to a lesser extent than in integral membrane proteins.5,9 The tight association can be rationalized by multivalent charge interactions between serum albumin and ligand.29 In fact, this protein contains a protected phospholipid-binding pocket, in addition to several low-affinity sites, in which the lipid is bound via contacts between basic residues and the ionic phosphate headgroup.30 Our data suggest that lipid interactions with selected binding sites on soluble proteins can mirror effects observed for membrane proteins.9
Figure 3.
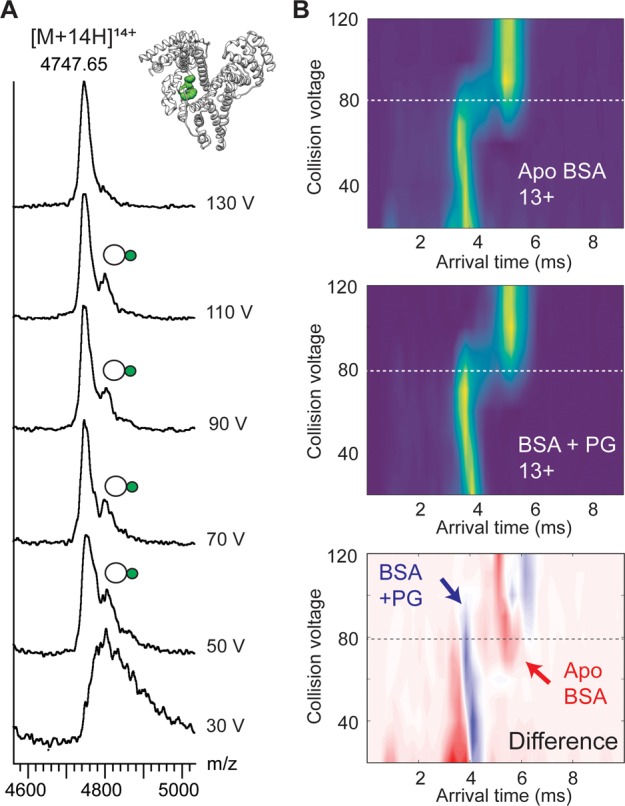
Lipid binding to BSA conveys gas-phase stabilization. (A) 1:1 complexes between BSA and POPG exhibit remarkable resistance to collisional dissociation. (Inset) Crystal structure of human serum albumin with lysophosphatidylethanolamine (green) bound to an internal binding site. (B) Collisional activation and ion mobility MS of POPG-bound BSA reveals higher resilience of the BSA–POPG complex to gas-phase unfolding compared to the apoprotein.
The fact that serum albumin–POPG complexes remain intact in the energy regime required for MS analysis of membrane proteins enabled us to compare the lipid-binding abilities of membrane proteins and soluble proteins directly. Serum albumin and the dimeric sodium-proton exchanger NapA, an integral membrane protein with no known lipid preferences,5,31 have comparable molecular masses (66 and 84 kDa, respectively) and retain bound lipids across a broad activation energy range. We therefore recorded mass spectra of a solution containing both proteins in 2× cmc C8E4 and a 10-fold excess of POPG at different collision voltages. Although an equimolar amount of both proteins was present, we found that only peaks assigned to serum albumin could be detected across the entire collision energy range (Figure 4). NapA, on the other hand, was detected only at collision voltages above 80 V. The higher activation energy required for observation of NapA is consistent with disruption of the detergent micelle prior to release into the gas phase and implies that NapA, but not serum albumin, is incorporated into detergent micelles.
Figure 4.
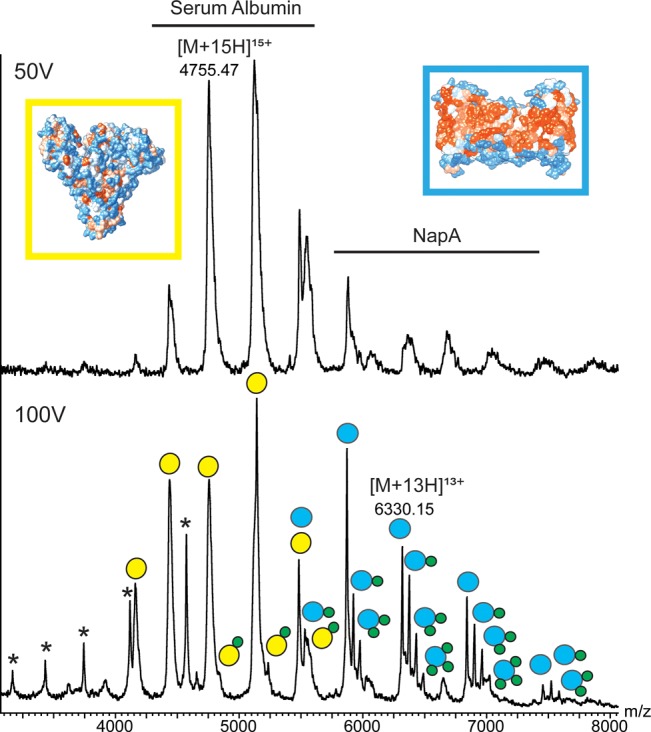
Simultaneous ESI-MS analysis of BSA and the integral membrane protein NapA. BSA can be detected at low collision voltages, suggesting that it is not incorporated into detergent micelles. Increasing the collision voltage releases NapA, revealing preferential binding of POPG. Asterisks indicate unfolded NapA monomers. POPG concentration is 25 μM.
The presence of a 10-fold excess of POPG leads to the appearance of multiple lipid adduct peaks of high intensity for NapA. This observation is in agreement with previous membrane protein studies.9,10 By contrast, only a single, minor lipid adduct peak was observed for serum albumin (Figure 4). Although the elevated collision voltage required to simultaneously observe free NapA and serum albumin has the potential to partially remove lipid adducts, these results clearly indicate that the integral membrane protein NapA has a much greater propensity to form protein–lipid complexes than the soluble serum albumin when both proteins are exposed to the same detergent/lipid environment. The preferential binding to NapA correlates well with the greater hydrophobic surface area within an overall comparable solvent-accessible surface (Table S1).
Discussion
In this study we investigated the factors that influence the association of lipids with soluble and membrane proteins in ESI-MS. While previous studies of soluble protein–lipid complexes focused on systems with specific lipid-binding abilities,32−34 we now show that the propensity of the common membrane lipids POPG, POPC, and POPE to form electrospray adducts is surprisingly low. The formation of protein–lipid complexes that can be preserved in the gas phase is instead critically dependent on the presence of detergent micelles in solution. We attribute this effect to the low cmc of phospholipids, which promotes the formation of lipid clusters in solution and thus limits their availability for association with proteins. In the presence of detergent, on the other hand, the negatively charged lipid POPG was found to associate readily with all proteins tested, while few, if any, complexes with zwitterionic lipids POPC or POPE could be detected.
When subjected to ESI-MS in the presence of detergent, the soluble proteins ubiquitin, MaSp1-NT, and BSA and the membrane protein NapA exhibit distinct lipid-binding properties: ubiquitin acquires low amounts of lipid adducts, in line with the notion that this protein does not engage in significant lipid interactions in vivo. Serum albumin, on the other hand, can tightly bind a single lipid molecule, suggesting a high-affinity binding site. MaSp1-NT exhibits a higher lipid-binding propensity than ubiquitin and BSA, suggesting that the availability of large charged and/or hydrophobic patches promotes association. Interestingly, soluble spidroins are stored in the sac of the silk gland alongside phospholipids.19 Molecular dynamics (MD) simulations suggest that the charged patches are freely accessible in the soluble proteins,19 and it may therefore be possible that lipids play a role in spidroin storage, perhaps by masking self-assembling protein interfaces, for example. The integral membrane protein NapA binds a large number of lipids with high resistance to collisional activation, as expected from its extensive interactions with the surrounding membrane.
Our results allow us to propose a general mechanism for the generation of protein–lipid complexes that can be observed in ESI-MS (Figure 5). Lipids preferentially incorporate into detergent micelles that provide shielding from the hydrophilic environment. During desolvation, the mixed lipid/detergent clusters dissociate, while lipids that bind with higher affinity than the detergent, for example via headgroup interactions, are left attached to the protein surface. Protein–lipid complexes are then formed via interactions between the proteins and the lipid-containing detergent micelles. Lipid binding to soluble proteins is accordingly promoted by increasing the concentration of detergent/lipid micelles. The ESI process favors nonspecific associations if multiple components are present in the same electrospray droplet, which has been estimated to arise at component concentrations above 50 μM.35 This is in agreement with our observation that lipid binding increases significantly at micelle concentrations above 200 μM. In detergents like DDM and C12E9, the low number of micelles (<5 μM at 2× cmc) limits the amount of ESI droplets that contain both protein and mixed detergent/lipid micelles, and virtually no protein–lipid complexes can be observed. Membrane proteins, on the other hand, are fully inserted into detergent micelles, exposing large surfaces for interactions with the embedded lipids, and complex formation occurs therefore in solution, independently of the electrospray process. Here, the entire protein–micelle complex is ionized as a single component instead. The detergent is subsequently removed by additional collisional activation, resulting in multiple lipids attached to the protein surface that was previously buried in the micelle.
Figure 5.
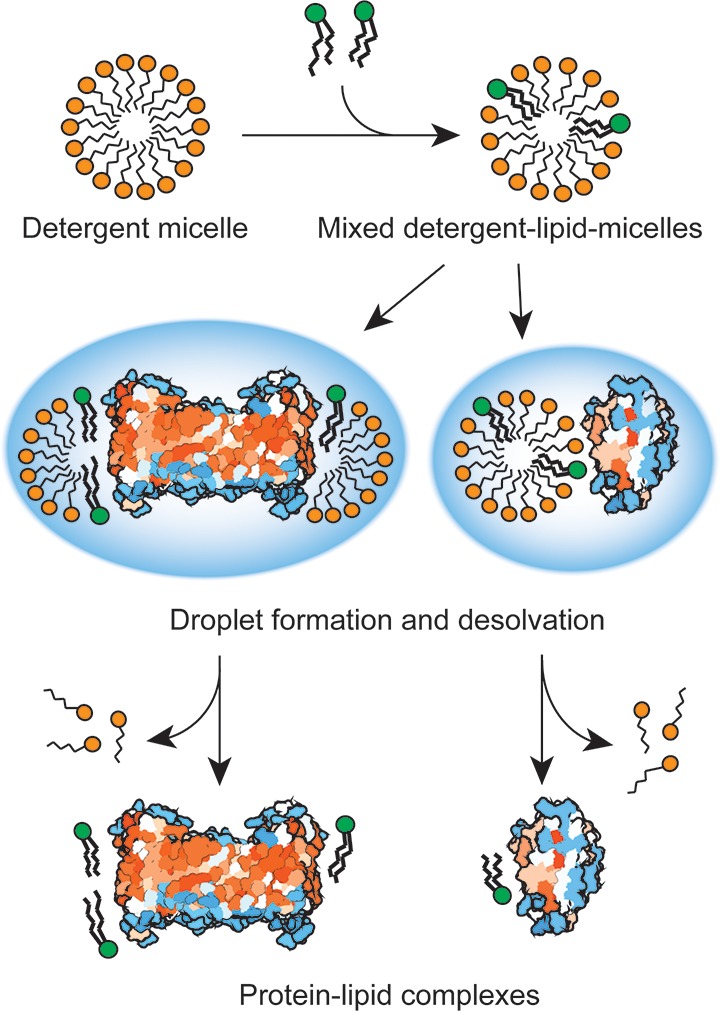
Mechanism for detergent-mediated formation of protein–lipid complexes. Lipids preferentially insert into detergent micelles. At sufficient micelle concentrations, soluble proteins and detergent/lipid micelles are ionized in the same electrospray droplet, making superficial contacts with the embedded lipids. Membrane proteins continually expose large surfaces to the intramicellar environment regardless of micelle concentration, leading to multiple lipid contacts. Desolvation and collisional activation remove the loosely bound detergent, releasing the protein–lipid complex for MS detection.
In summary, we show that ESI-MS captures interactions between membrane proteins and exogenous lipids as they occur inside detergent micelles, while the contributions from nonspecific electrospray adduct formation are negligible. ESI-MS is therefore suitable to study even transient protein–lipid interactions that may be inaccessible by other approaches.
Acknowledgments
We gratefully acknowledge helpful discussions with the group members of C.V.R.’s laboratory. Professor Jan Johansson and Professor Anna Rising, Department of Neurobiology, Care Sciences and Society (NVS), Karolinska Institutet, Stockholm, are thanked for the MaSp1-NT protein and comments on the manuscript, and Professor David Drew, Department of Biochemistry and Biophysics, Stockholm University, is thanked for the NapA protein and helpful discussions. M.L. holds an ERC Marie Curie early career development grant and is a junior research fellow of St. Cross College, University of Oxford. E.G.M. holds a Marie Skłodowska Curie international career grant from the European Commission and the Swedish Research Council and is a Fulford junior research fellow at Somerville College, University of Oxford. J.G. is a junior research fellow of The Queen’s College, University of Oxford, and is supported by Wellcome Trust grant 104633/Z/14/Z held by C.V.R. C.V.R. is supported by an ERC advanced investigator award 695511 (ENABLE), a Wellcome Trust investigator award (104633/Z/14/Z), and a Medical Research Council Program grant (MR/N020413/1).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b00922.
One table listing hydrophobic, hydrophilic, and total solvent-accessible surface areas and number of charged residues of proteins; four figures showing release of NapA from DDM and C8E4 micelles, differential lipid binding preferences, detection of free lipids by ESI-MS, and disruption of complexes between MaSP1-NT and POPG (PDF)
Author Present Address
§ Department of Microbiology, Tumour and Cell Biology, Karolinska Institutet and Science for Life Laboratory, SE-171 65 Stockholm, Sweden (M.L.).
The authors declare no competing financial interest.
Supplementary Material
References
- Barrera N. P.; Di Bartolo N.; Booth P. J.; Robinson C. V. Science 2008, 321, 243–246. 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- Barrera N. P.; Zhou M.; Robinson C. V. Trends Cell Biol. 2013, 23, 1–8. 10.1016/j.tcb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Bechara C.; Robinson C. V. J. Am. Chem. Soc. 2015, 137, 5240–5247. 10.1021/jacs.5b00420. [DOI] [PubMed] [Google Scholar]
- Gupta K.; Donlan J. A.; Hopper J. T.; Uzdavinys P.; Landreh M.; Struwe W. B.; Drew D.; Baldwin A. J.; Stansfeld P. J.; Robinson C. V. Nature 2017, 541, 421–424. 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreh M.; Marklund E. G.; Uzdavinys P.; Degiacomi M. T.; Coincon M.; Gault J.; Gupta K.; Liko I.; Benesch J. L.; Drew D.; Robinson C. V. Nat. Commun. 2017, 8, 13993. 10.1038/ncomms13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara C.; Noll A.; Morgner N.; Degiacomi M. T.; Tampe R.; Robinson C. V. Nat. Chem. 2015, 7, 255–262. 10.1038/nchem.2172. [DOI] [PubMed] [Google Scholar]
- Demmers J. A.; van Dalen A.; de Kruijff B.; Heck A. J.; Killian J. A. FEBS Lett. 2003, 541, 28–32. 10.1016/S0014-5793(03)00282-5. [DOI] [PubMed] [Google Scholar]
- Pliotas C.; Dahl A. C.; Rasmussen T.; Mahendran K. R.; Smith T. K.; Marius P.; Gault J.; Banda T.; Rasmussen A.; Miller S.; Robinson C. V.; Bayley H.; Sansom M. S.; Booth I. R.; Naismith J. H. Nat. Struct. Mol. Biol. 2015, 22, 991–998. 10.1038/nsmb.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A.; Reading E.; Allison T. M.; Ulmschneider M. B.; Degiacomi M. T.; Baldwin A. J.; Robinson C. V. Nature 2014, 510, 172–175. 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux J.; Wang S. C.; Politis A.; Reading E.; Ma J.; Biggin P. C.; Zhou M.; Tao H.; Zhang Q.; Chang G.; Morgner N.; Robinson C. V. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 9704–9709. 10.1073/pnas.1303888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C.; Stein R. A.; Masureel M.; Roth A.; Mishra S.; Dawaliby R.; Konijnenberg A.; Sobott F.; Govaerts C.; McHaourab H. S. Nat. Struct. Mol. Biol. 2016, 23, 744–751. 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Kitova E. N.; Klassen J. S. Anal. Chem. 2003, 75, 4945–4955. 10.1021/ac034300l. [DOI] [PubMed] [Google Scholar]
- Robinson C. V.; Chung E. W.; Kragelund B. B.; Knudsen J.; Aplin R. T.; Poulsen F. M.; Dobson C. M. J. Am. Chem. Soc. 1996, 118, 8646–8653. 10.1021/ja960211x. [DOI] [Google Scholar]
- Wang W.; Kitova E. N.; Klassen J. S. Anal. Chem. 2005, 77, 3060–3071. 10.1021/ac048433y. [DOI] [PubMed] [Google Scholar]
- McAllister R. G.; Metwally H.; Sun Y.; Konermann L. J. Am. Chem. Soc. 2015, 137, 12667–12676. 10.1021/jacs.5b07913. [DOI] [PubMed] [Google Scholar]
- Charbonneau D. M.; Tajmir-Riahi H. A. J. Phys. Chem. B 2010, 114, 1148–1155. 10.1021/jp910077h. [DOI] [PubMed] [Google Scholar]
- Petitpas I.; Grune T.; Bhattacharya A. A.; Curry S. J. Mol. Biol. 2001, 314, 955–960. 10.1006/jmbi.2000.5208. [DOI] [PubMed] [Google Scholar]
- Askarieh G.; Hedhammar M.; Nordling K.; Saenz A.; Casals C.; Rising A.; Johansson J.; Knight S. D. Nature 2010, 465, 236–238. 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- dos Santos-Pinto J. R.; Arcuri H. A.; Priewalder H.; Salles H. C.; Palma M. S.; Lubec G. J. Proteome Res. 2015, 14, 3859–3870. 10.1021/acs.jproteome.5b00243. [DOI] [PubMed] [Google Scholar]
- Kronqvist N.; Otikovs M.; Chmyrov V.; Chen G.; Andersson M.; Nordling K.; Landreh M.; Sarr M.; Jornvall H.; Wennmalm S.; Widengren J.; Meng Q.; Rising A.; Otzen D.; Knight S. D.; Jaudzems K.; Johansson J. Nat. Commun. 2014, 5, 3254 10.1038/ncomms4254. [DOI] [PubMed] [Google Scholar]
- le Maire M.; Champeil P.; Møller J. V. Biochim. Biophys. Acta, Biomembr. 2000, 1508, 86–111. 10.1016/S0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Allison T. M.; Reading E.; Liko I.; Baldwin A. J.; Laganowsky A.; Robinson C. V. Nat. Commun. 2015, 6, 8551. 10.1038/ncomms9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobott F.; Hernandez H.; McCammon M. G.; Tito M. A.; Robinson C. V. Anal. Chem. 2002, 74, 1402–1407. 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- Loo J. A. Mass Spectrom. Rev. 1997, 16, 1–23. . [DOI] [PubMed] [Google Scholar]
- Laganowsky A.; Reading E.; Hopper J. T.; Robinson C. V. Nat. Protoc. 2013, 8, 639–651. 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading E.; Liko I.; Allison T. M.; Benesch J. L.; Laganowsky A.; Robinson C. V. Angew. Chem., Int. Ed. 2015, 54, 4577–4581. 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- Landreh M.; Alvelius G.; Johansson J.; Jornvall H. Anal. Chem. 2014, 86, 4135–4139. 10.1021/ac500879c. [DOI] [PubMed] [Google Scholar]
- Hopper J. T.; Oldham N. J. J. Am. Soc. Mass Spectrom. 2009, 20, 1851–1858. 10.1016/j.jasms.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Bartman C. E.; Metwally H.; Konermann L. Anal. Chem. 2016, 88, 6905–6913. 10.1021/acs.analchem.6b01627. [DOI] [PubMed] [Google Scholar]
- Guo S.; Shi X.; Yang F.; Chen L.; Meehan E. J.; Bian C.; Huang M. Biochem. J. 2009, 423, 23–30. 10.1042/BJ20090913. [DOI] [PubMed] [Google Scholar]
- Lee C.; Kang H. J.; von Ballmoos C.; Newstead S.; Uzdavinys P.; Dotson D. L.; Iwata S.; Beckstein O.; Cameron A. D.; Drew D. Nature 2013, 501, 573–577. 10.1038/nature12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson C. L.; Ilag L. L.; Malo J.; Hatters D. M.; Howlett G. J.; Robinson C. V. Biophys. J. 2003, 85, 3802–3812. 10.1016/S0006-3495(03)74795-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brouwer A. P.; Versluis C.; Westerman J.; Roelofsen B.; Heck A. J.; Wirtz K. W. Biochemistry 2002, 41, 8013–8018. 10.1021/bi016055a. [DOI] [PubMed] [Google Scholar]
- Liu L.; Bagal D.; Kitova E. N.; Schnier P. D.; Klassen J. S. J. Am. Chem. Soc. 2009, 131, 15980–15981. 10.1021/ja9060454. [DOI] [PubMed] [Google Scholar]
- Benesch J. L.; Ruotolo B. T.; Simmons D. A.; Robinson C. V. Chem. Rev. 2007, 107, 3544–3567. 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


