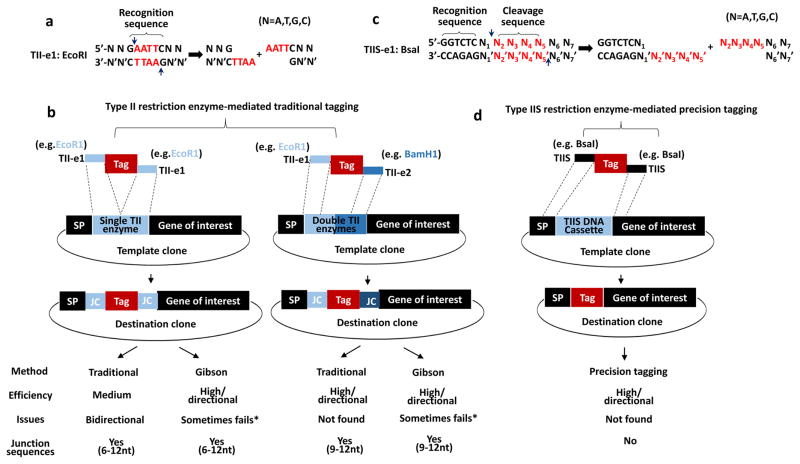
Figure 2. Comparison of type II and IIS restriction enzyme-mediated protein tagging strategy.
(a) Type II restriction enzymes (TII-es) recognize palindromic DNA sequences. For example, EcoRI recognizes 5′-GAATTC-3′ (marked by top curly bracket) and creates 4 base pairs overhangs highlighted in red.
(b) Single or double type II restriction enzymes cassette (highlighted in blue box) for traditional protein tagging. Note that in all destination clones, varying junction sequences exist adjacent to both sides of the tag.
(c) Type IIS restriction enzymes (TIIS-es) recognize non-palindromic, asymmetrical DNA sequences. For example, BsaI recognizes 5′-GGTCTC-3′ (marked by top curly bracket) and cleaves DNA one bp away (indicated by the two arrows), producing N2′N3′N4′N5′ custom sticky end (highlighted in red). N indicates four bases of DNA, including A, T, G and C. Apostrophe (’) indicates the complementary base of the DNA.
(d) Type IIS restriction enzyme DNA cassette (TIIS DNA cassette highlighted in blue box) for precision tagging. Note that on both ends of a tag, the flanking sequences (such as BsaI-released 5′-N2′N3′N4′N5′ and 5′-N2N3N4N5 belong to gene-specific sequences including SP or gene of interest indicated by two closely dotted lines. After Tag replaces type IIS DNA cassette, a scarless tagging clone can be generated. Comparison between traditional and precision tagging were summarized in the bottom table. * Gibson assembly sometimes fails due to certain DNA sequences such as repetitive region or creating one or two nucleotides deletion.