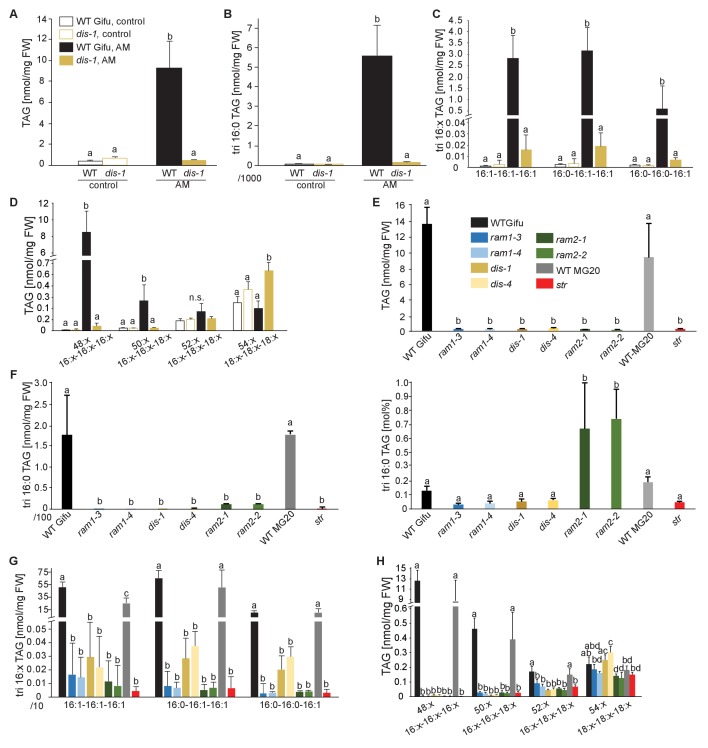
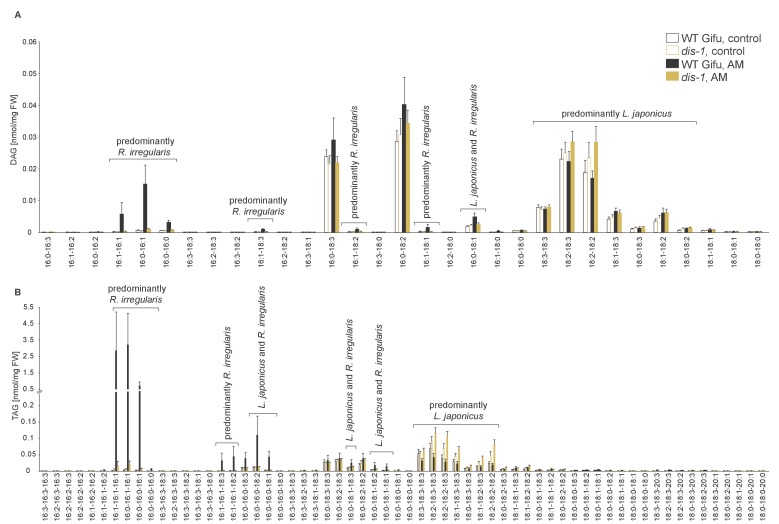
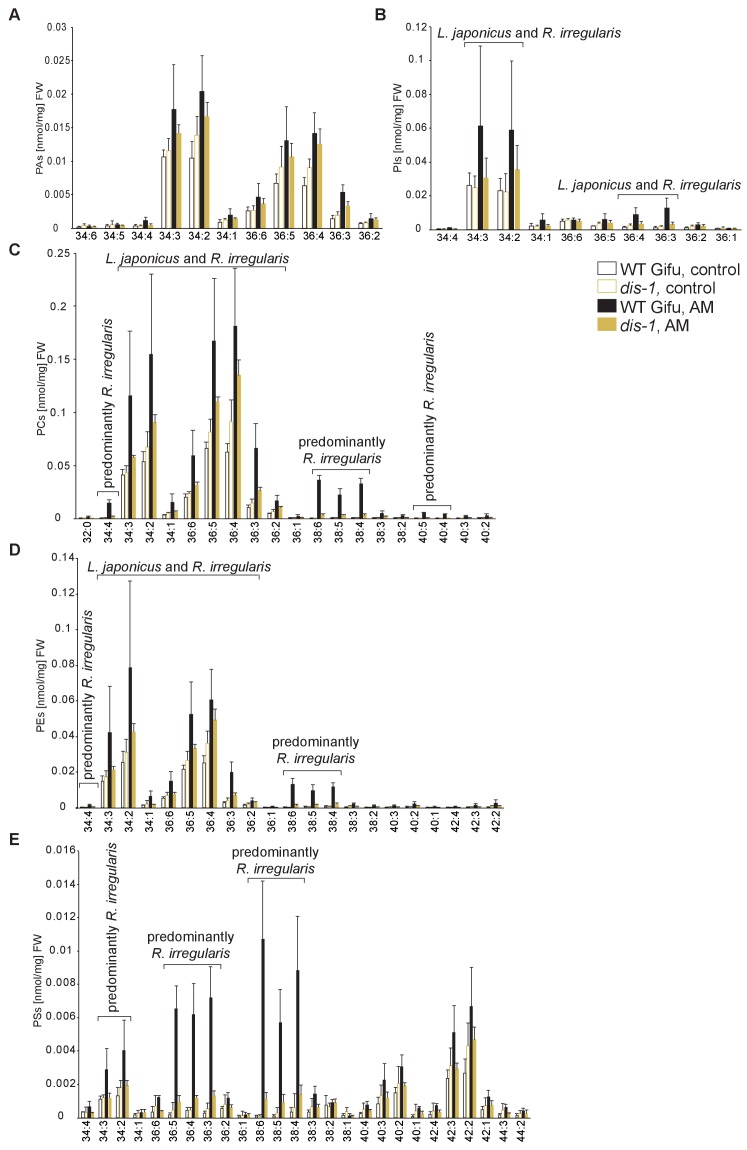
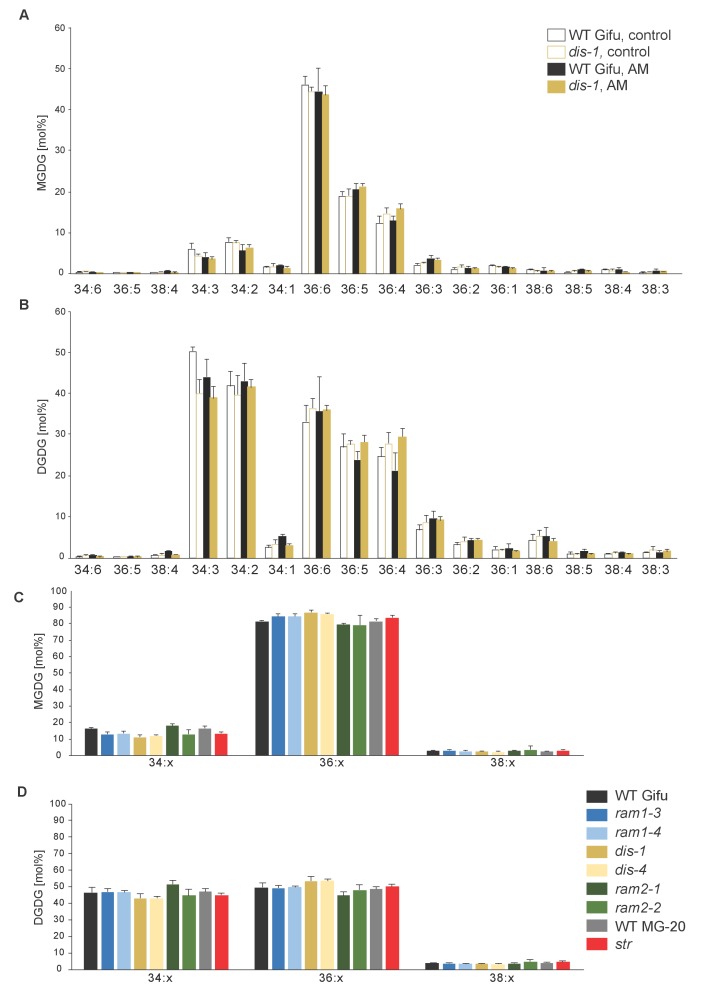
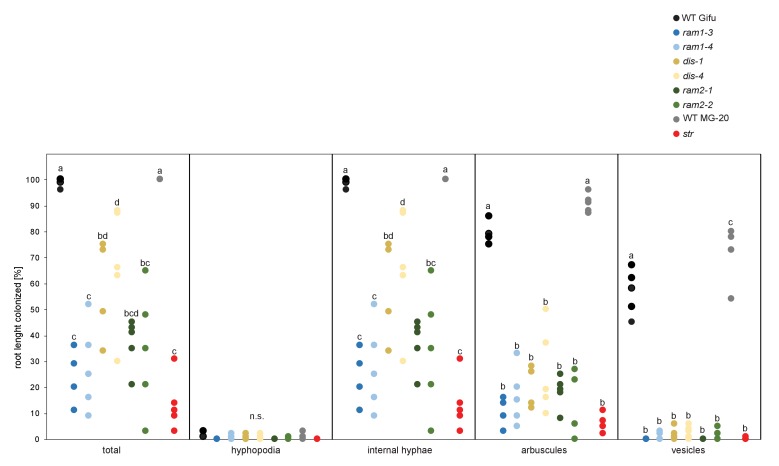
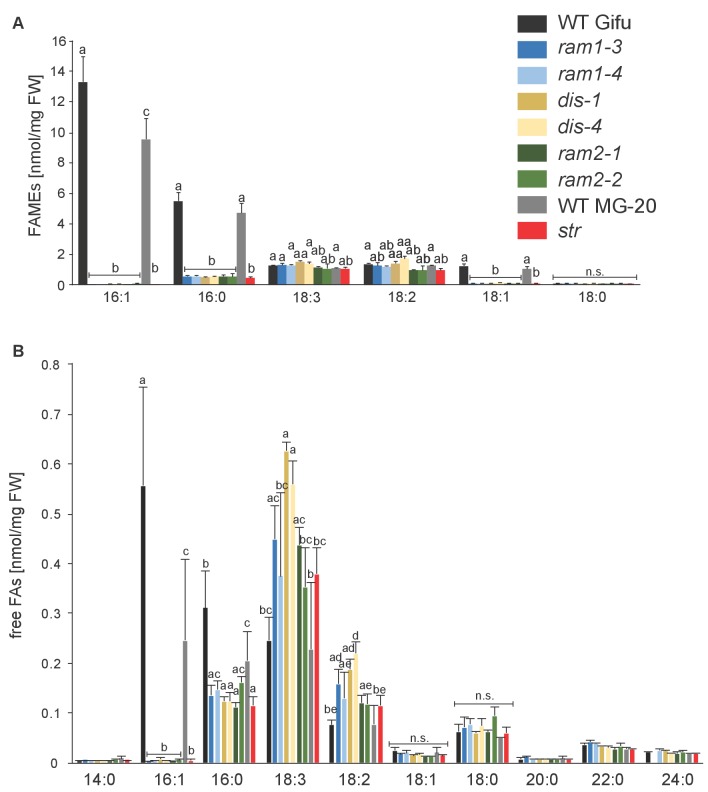
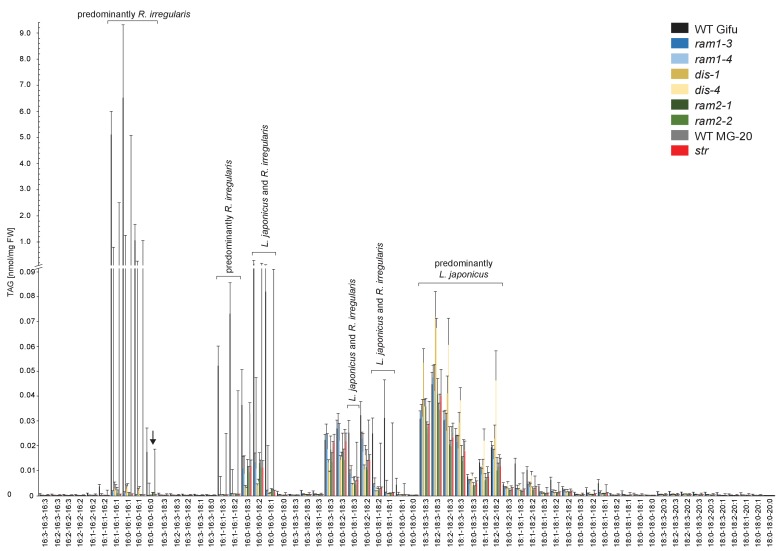
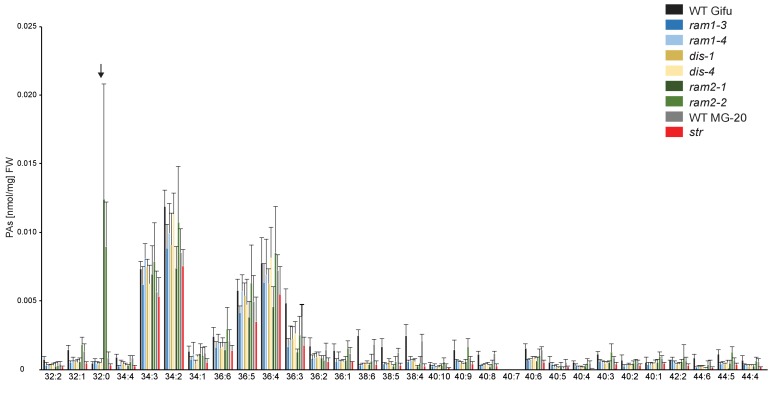
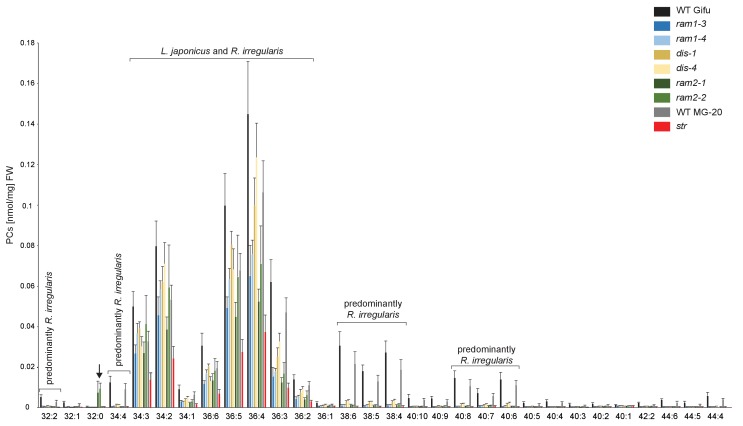
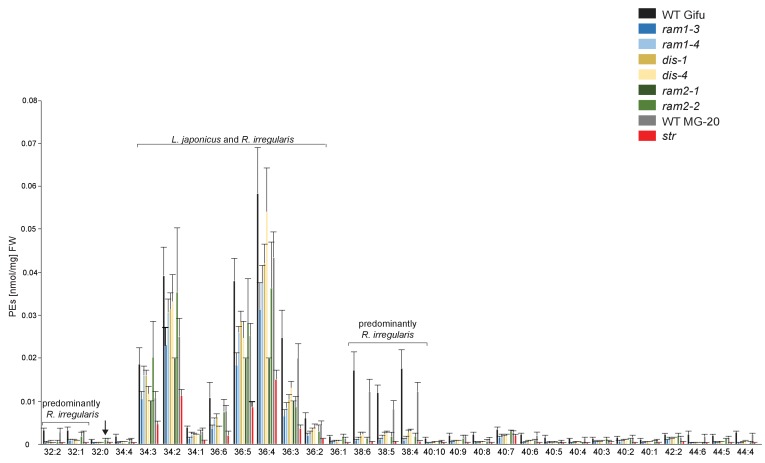
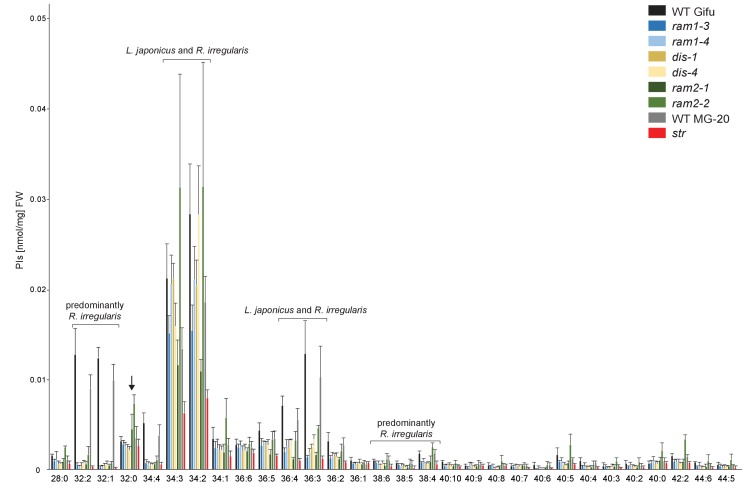
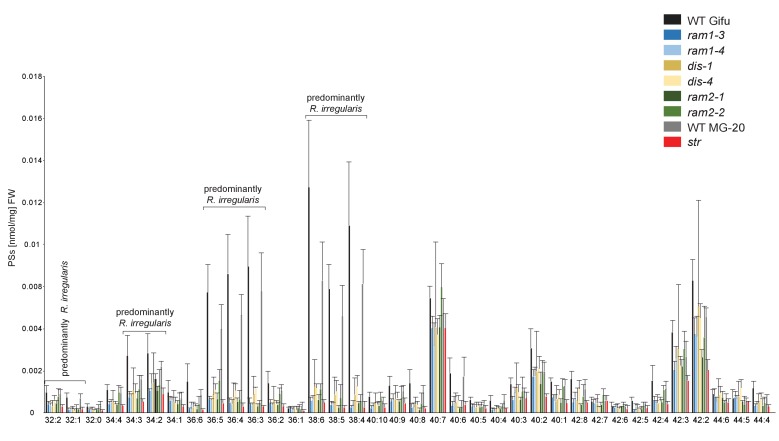
Figure 5. Lack of characteristic accumulation of triacylglycerols in AM-defective mutants.
(A-D) Quantitative accumulation of (A) total triacylglycerols, (B) tri16:0-triacylglycerol (C) tri16:x-triacylglycerols and (D) of triacylglycerols harbouring 16:x and 18:x FA-chains in non-colonized and R. irregularis colonized wild-type and dis-1 roots. Different letters indicate significant differences (ANOVA; posthoc Tukey) (A): n = 18; p≤0.001; F3,14 = 68.16. (B): n = 18; p≤0.001; F3,14 = 68.48. (C): n = 19; p≤0.01, F3,15 = 7.851 (16:1-16:1-16:1); p≤0.001, F3,15 = 14.52 (16:0-16:1-16:1); p≤0.001, F3,15 = 39.22 (16:0-16:0-16:1). (D): n = 19; p≤0.001, F3,15 = 12.15 (48:x), F3,15 = 15.56 (50:x); p≤0.01, F3,15 = 22.93 (54:x). (E-G) Quantitative accumulation of (E) total triacylglycerols, (F) tri16:0-triacylglycerols, (G) tri16:x-triacylglycerols and (H) of triacylglycerols harbouring 16:x and 18:x FA-chains in colonized roots of L. japonicus wild-type Gifu, wild-type MG-20 and arbuscule-defective mutants. Different letters indicate significant differences (ANOVA; posthoc Tukey). (E): n = 40; p≤0.001; F8,31 = 38.42. (F) Left: absolute tri16:0 TAG content: n = 40; p≤0.001; F8,31 = 19.05. Right: tri16:0 TAG proportion among all TAGs, n = 40; p≤0.001; F8,31 = 14.21. (G): p≤0.001; n = 41, F8,32 = 86.16 (16:1-16:1-16:1); n = 39, F8,30 = 24.16 (16:0-16:1-16:1); n = 40, F8,31 = 17.67 (16:0-16:0-16:1). (H): n = 40; p≤0.001, F8,31 = 39.26 (48:x), F8,31 = 28.93 (50:x); p≤0.01, F8,31 = 19.78 (52:x); p≤0.05, F8,31 = 13.77 (54:x). (A-H) Bars represent means ±standard deviation (SD) of 3–5 biological replicates.
DOI: http://dx.doi.org/10.7554/eLife.29107.022