Fig. 2. Effect of drug treatment on primary tumor transcriptome indicators of EMT, tumor-associated leukocytes, and pro-metastatic transcription factors.
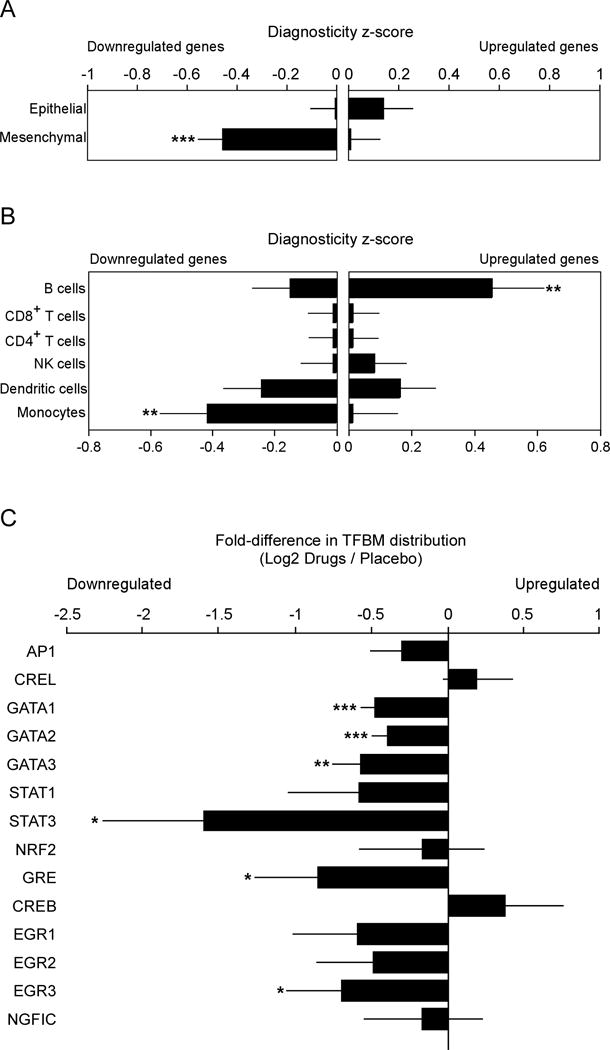
Twenty-five tumors yielded RNA of sufficient quality for transcriptome profiling (10 drug-treated and 15 placebo). (A) Effects of drug treatment on primary tumor EMT gene expression were quantified by Transcript Origin Analysis (58) of 163 genes showing > 1.25-fold up-regulation and 141 genes showing equivalent down-regulation in tumors from drug-treated patients vs. controls, using reference transcriptome profiles derived from mesenchymal- vs. epithelial-polarized breast cancer cells (59). (B) Transcript Origin Analysis also assessed the effects of drug treatment on expression of genes derived from monocytes, dendritic cells, CD4+ and CD8+ T cells, B cells and NK cells, using reference data derived from isolated samples of each cell type (22). (C) Effect of drug treatment on transcription control pathways as indicated by bioinformatics analysis of transcription factor-binding motifs in promoters of differentially expressed genes. Data is presented as mean ± SEM. Group differences are indicated by * (p < 0.05), ** (p < 0.01), or *** (p < 0.001).
