Figure 4.
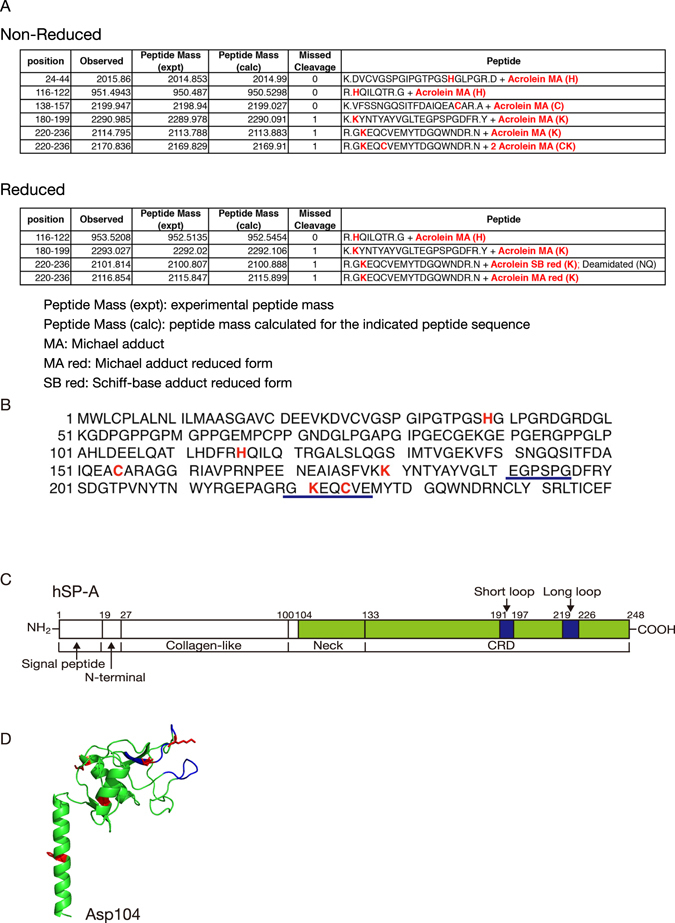
The identification of acrolein-modified residues in SP-A. (A) Nano-LC/MALDI-TOF MS/MS analysis of peptides obtained from the trypsin digestion of acrolein-modified hSP-A. Trypsin digested acrolein-modified SP-A was subjected to Nano-LC/MALDI-TOF MS/MS analysis and identified by using MASCOT Daemon. Acrolein-modified residues are shown in red. (Upper) The MS/MS analysis of non-reduced acrolein-modified SP-A. (Lower) The MS/MS analysis of NaBH4-reduced acrolein-modified SP-A. (B) The amino acid sequence of hSP-A. Acrolein-modified residues are shown in red, and the double-loop structure is indicated by the blue underlined text. (C) The structural organisation of human SP-A. SP-A is conceptually divided into a signal peptide, N-terminal, collagen-like neck and CRD domain. The double-loop structure in the CRD domain is shown in blue (long loop residues: 219–226 and short loop residues: 191–197). Green corresponds to residues 104–248, which are shown in the crystal structure in (D). (D) A homology model of the hSP-A neck and CRD domains (residues: 104–248) based on the crystal structure of rat SP-A (PDB ID: 1R13). The model was constructed using SWISS-MODEL37. The protein backbone is shown in green, the double-loop structure in the CRD domain is shown in blue, and acrolein-modified residues are highlighted in red.
