Figure 3.
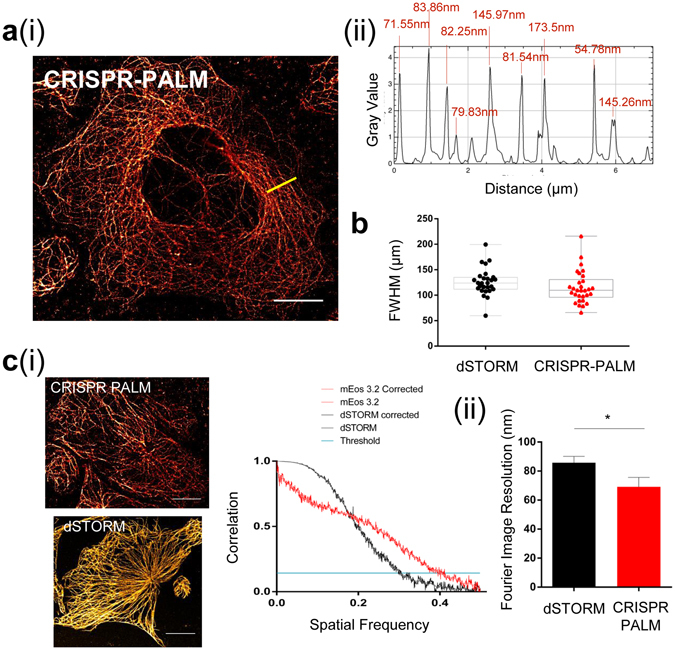
Genomic expression of a tagged TubA1B allows for SMLM equivalent to dSTORM images. (a) (i) PALM imaging of CRISPR mEos 3.2 results in clean, consistent super-resolved tubulin fibers despite the labelling density required for high quality cytoskeletal imaging. (ii) shows an example FWHM line profile for the yellow line in (a) (i). (b) Median Full width half maxima (FWHM) of line profiles across multiple cells shows that mEos 3.2 FWHM in Hel 92.1.7 cells is not significantly different when compared to dSTORM (median 123.91 (dSTORM) and 109.713 (mEos 3.2)) (c) (i) Applying the Fourier Ring Correlation method also reveals Fourier Image Resolution (FIRE) values in xy of 40 nm for mEos 3.2 and 72 nm for dSTORM representative figures. FRC plots for the representative images are shown from which is calculated the FIRE value. (ii) When measured across all data sets, a statistically significant improvement in FIRE is observed (*p = 0.0447). FWHM data generated from line profiles in Fiji, with measurements obtained from multi-peak analysis in Igor Pro. Box and whisker plot showing median with min and max values. FIRE values and FRC curves are calculated using FRC plug-in (Fiji), with an unpaired t-test used to test for significance. (10 μm scale).
