Fig. 4.
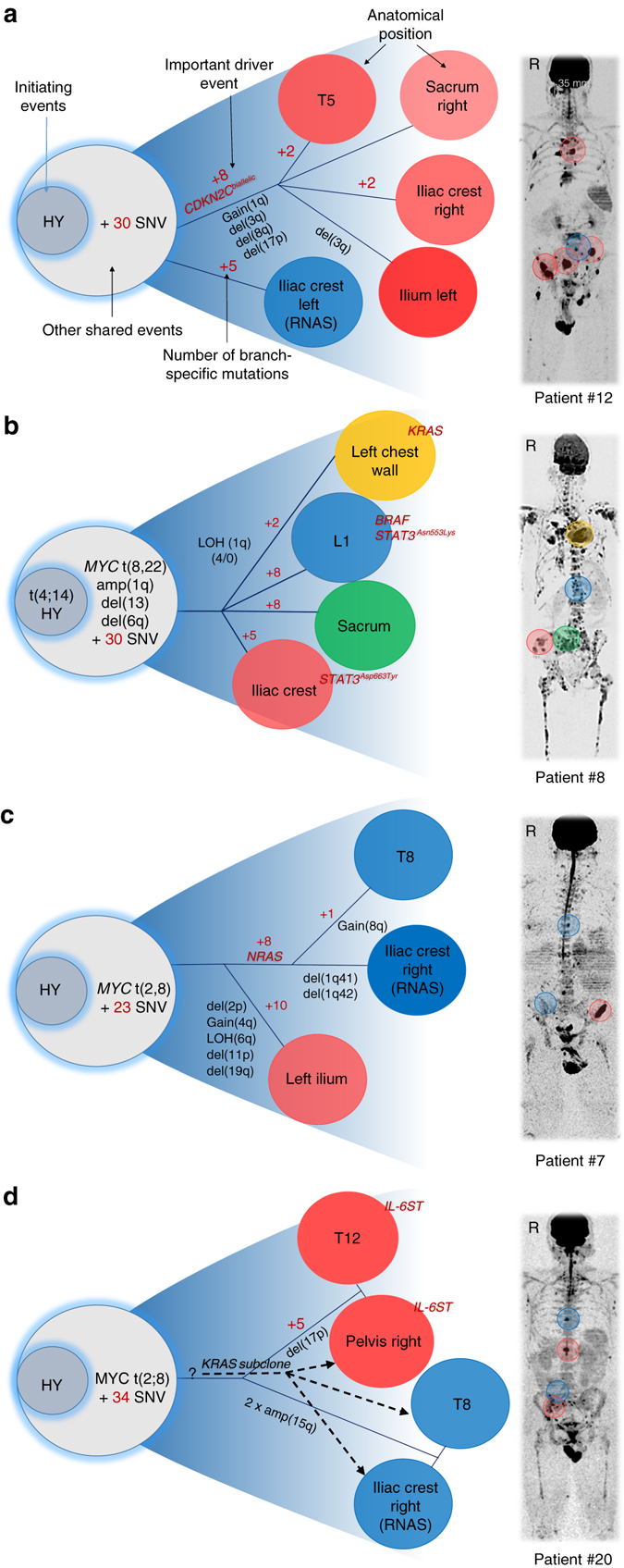
Multi-regional evolution. Cases with availability of multiple CT-guided samples were selected to analyze the phylogenetic relationship of clones from different regions. The location of samples is marked in the medical images in the right panel using the color code that was assigned to clones (left panel). The letter R indicates the right side of the body. a Four focal lesions (FL) showed similar genomic profiles, sharing the high-risk events bi-allelic deletion of CDKN2C, gain(1q) and del(17p). In contrast, a randomly biopsied non-FL site was low-risk and showed none of these events. b FLs displayed different genomic profiles with each of them containing unique driver mutations (BRAF, KRAS, and STAT3). Clones at the iliac crest and L1 contained different STAT3 mutations (STAT3Asn553Lys vs. STAT3Asp663Tyr). c Clones at T8 and the left iliac crest shared a missense NRAS mutation. Compared to the NRAS branch, the right iliac crest FL showed multiple site-specific CNAs and non-silent mutations. d Two major clones occupied four distant sites with the first clone being characterized by a non-ubiquitous IL6ST mutation. Moreover, an independent minor subclone with a missense KRAS mutation infiltrated both sites in the pelvis and T8, but not T12
