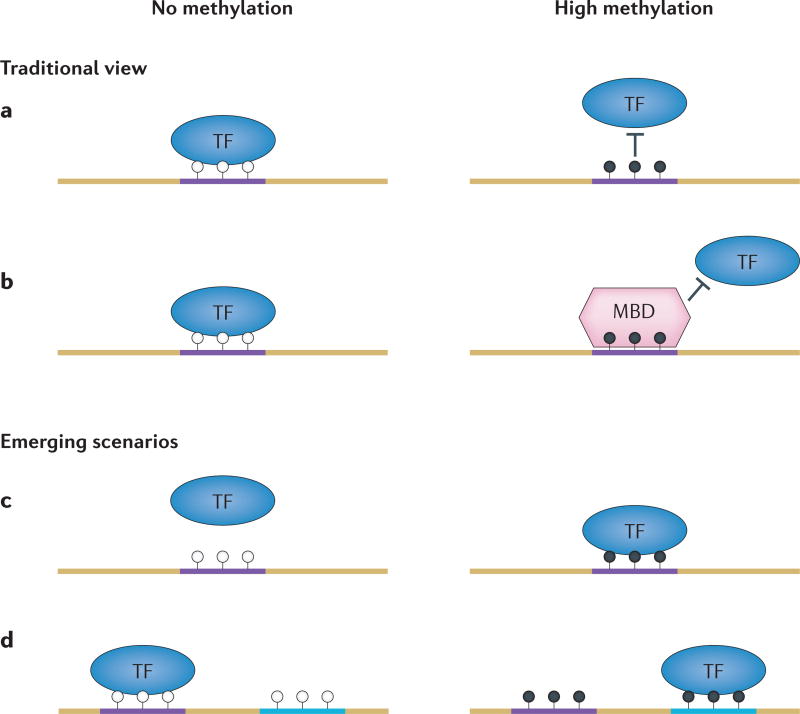
Figure 1. Interaction modes between proteins and DNA.
a,b | The traditional view of the protein–DNA interaction patterns. Transcription factors (TFs) usually bind to non-methylated DNA motifs (open circles, left panels) in open chromatin regions (part a). However, such interactions can be directly disrupted by methylation on the CpG sites in the motifs (filled circles, right panels). Alternatively, methyl-CpG binding-domain (MBD) proteins can be recruited to the methylated DNA motifs and compete off TFs through their higher affinity to the mCpG site in a sequence-independent fashion (part b). c,d | Newly emerging scenarios for protein–DNA interactions. DNA methylation could create a new binding site for TFs (part c). TFs may be able to recognize different sequences with or without DNA methylation (part d).