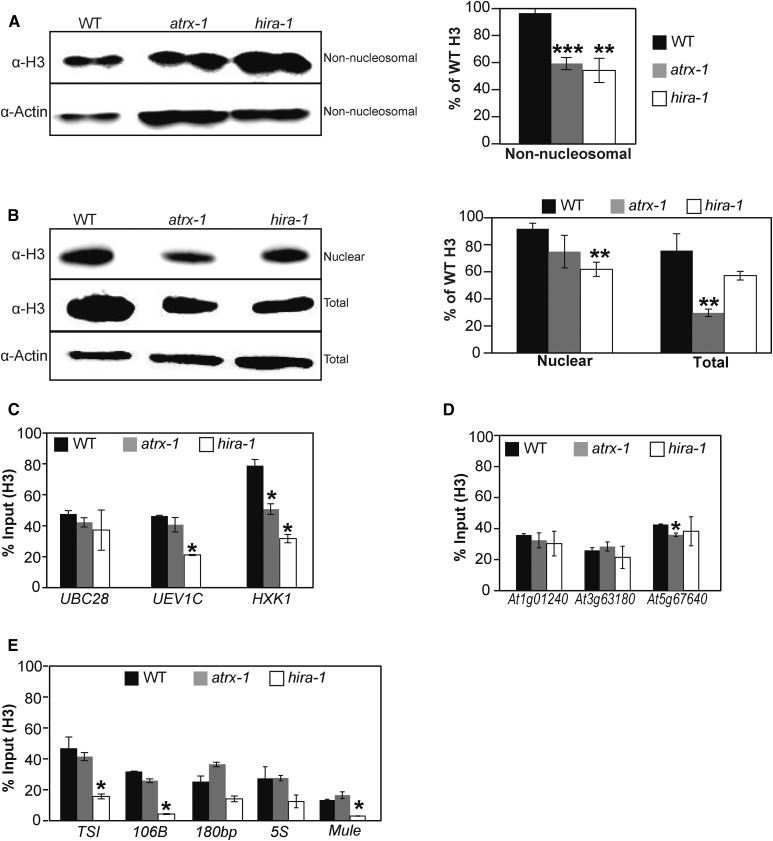
Figure 4.
Effects of ATRX Loss on Histone Pools and Nucleosome Occupancy.
(A) and (B) Left: Histone H3 protein levels quantified by immunoblot in non-nucleosomal fractions (A) and nuclear and total extracts (B). Twenty (A) or six (B) micrograms of proteins were loaded per lane. Right: Quantification of H3 band intensities normalized to Actin from two independent experiments, each comprising two biological replicates for each genotype, consisting of independent pools of 1 g of 2.5-week-old in vitro-grown plantlets collected at the same time, and several blots. Student’s t test compared with the wild type; **P < 0.01 and ***P < 0.001.
(C) to (E) Histone H3 occupancy at three active genes (UBC28, UEV1C, and HXK1) (C), at three genes situated in subtelomeric regions (At1g01240, At3g63180, and At5g67640) (D), and at centromeric and pericentromeric repeats (TSI, 106B, 180bp, and ribosomal 5S rDNA loci) and at a transposon on a chromosome arm (At2g15810, Mule) (E) was assessed by H3-ChIP qPCR in three biological replicates consisting of pools of 1 g of 2.5-week-old in vitro-grown wild-type, atrx-1, and hira-1 mutant plants. Student’s t test compared with the wild type; *P < 0.05.
Error bars of all panels represent se of the mean.