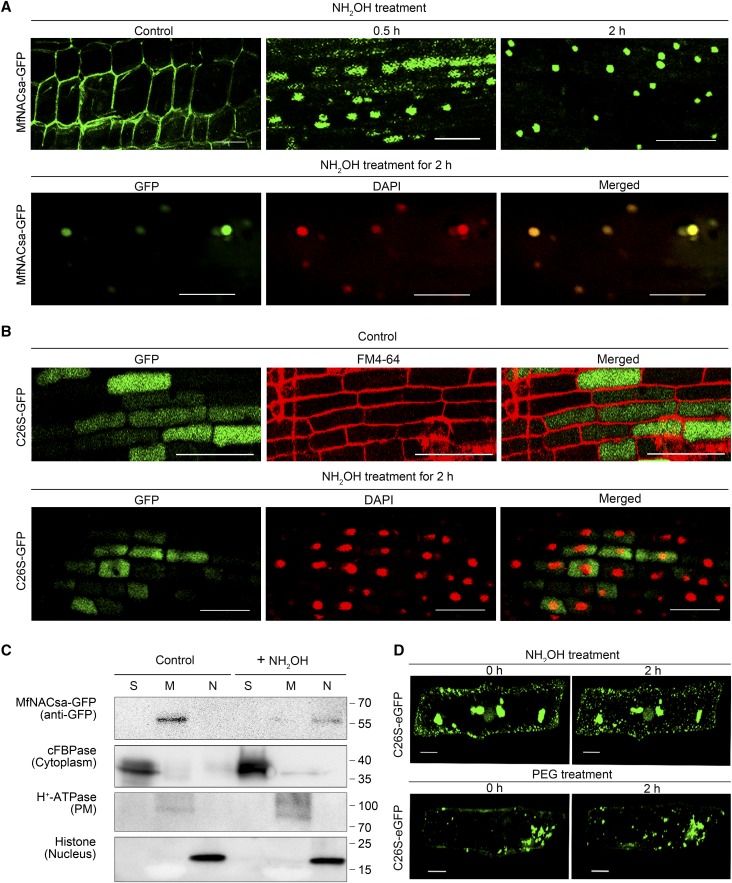
Figure 7.
MfNACsa Is Translocated to the Nucleus through De-S-Palmitoylation.
(A) Transgenic M. truncatula hairy roots expressing the Pro35S:MfNACsa-GFP fusion proteins were treated with 0.05 M hydroxylamine (NH2OH). The GFP signal pattern was detected by confocal imaging at 0 (control), 0.5, or 2 h after NH2OH treatment. After 0.05 M NH2OH treatment for 2 h, the nucleus was stained with DAPI (pseudocolor, red), with the ICQ of 0.436. Bars = 50 μm.
(B) Transgenic M. truncatula hairy roots expressing the Pro35S:C26S -GFP mutant proteins were treated with 0.05 M NH2OH. The GFP signal pattern was detected by confocal imaging (Olympus FluoView FV1000) under control conditions, and the negative (–0.037) ICQ of C26S-GFP against FM4-64. After 0.05 M NH2OH treatment for 2 h, the nucleus was stained with DAPI (pseudocolor, red) and the negative (–0.006) ICQ of C26S-GFP against DAPI dye. Bars = 50 μm.
(C) Cellular fraction analysis of transient transgenic N. benthamiana leaves expressing MfNACsa-GFP proteins at 0 and 2 h after NH2OH treatment. Presence of MfNACsa-GFP was shown. cFBPase, H+-ATPase, and Histone H3 are used as cytoplasm, PM, and nuclear fraction markers, respectively.
(D) Onion epidermal cells expressing C26S-eGFP mutant proteins were treated with 0.05 M NH2OH or 10% PEG-8000 for 2 h, respectively. The signal patterns were observed by fluorescence microscopy (Olympus FluoView FV1000). Bars = 50 μm.