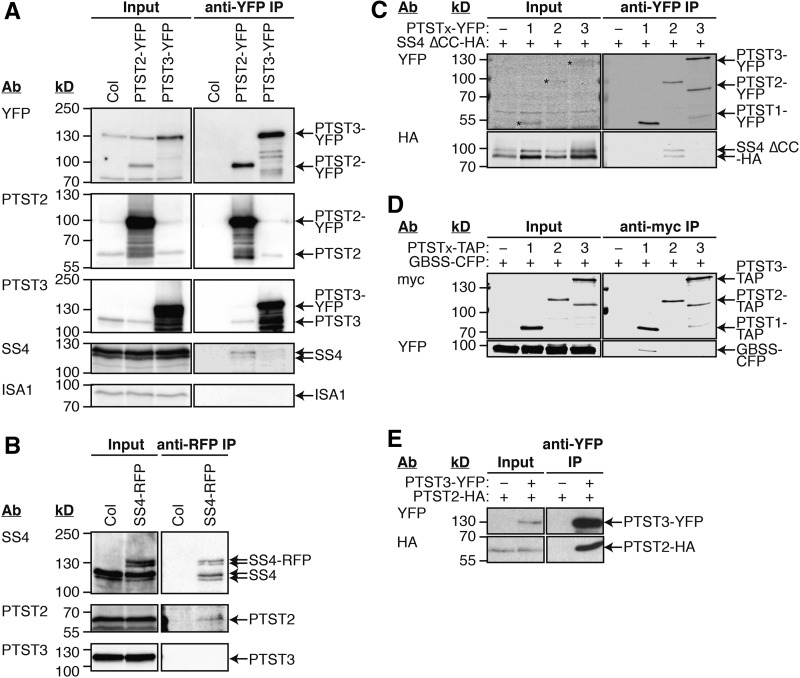
Figure 8.
Immunoprecipitation Experiments for Interaction Partners of PTST2 and PTST3.
The antibodies (Ab) used for detection and the migration of molecular mass markers (in kD) are indicated on the left of all immunoblot panels.
(A) Immunoprecipitation of PTST2-YFP and PTST3-YFP expressed in Arabidopsis. Proteins extracted from leaf tissue (Input) were incubated with beads conjugated to YFP antibodies. The bound proteins were eluted (anti-YFP IP), and proteins in the input and IP fractions were detected by immunoblotting.
(B) Immunoprecipitation of SS4-RFP expressed in Arabidopsis. The experiment was conducted as for (A), except that anti-RFP beads were used. SS4, PTST2, and PTST3 proteins in the input and IP fractions were detected by immunoblotting.
(C) Immunoprecipitation of YFP-tagged PTST proteins coexpressed in N. benthamiana leaves with HA-tagged SS4 protein. A truncated version of SS4 lacking the N-terminal coiled coils (SS4 ∆CC) was used. Immunoprecipitation was conducted as for (A). Epitope-tagged proteins in the Input and IP fractions were detected using immunoblotting.
(D) Immunoprecipitation of PTST proteins tagged with TAP-tags, coexpressed in N. benthamiana leaves with CFP-tagged GBSS protein. The experiment was conducted as described for (C), except using anti-myc beads, which bind the c-myc epitopes in the TAP tag.
(E) Immunoprecipitation of YFP-tagged PTST3 coexpressed in N. benthamiana leaves with HA-tagged PTST2 protein. The experiment was conducted as described for (A). Epitope-tagged proteins in the input and IP fractions were detected using anti-YFP (upper panels) and anti-HA (lower panels).