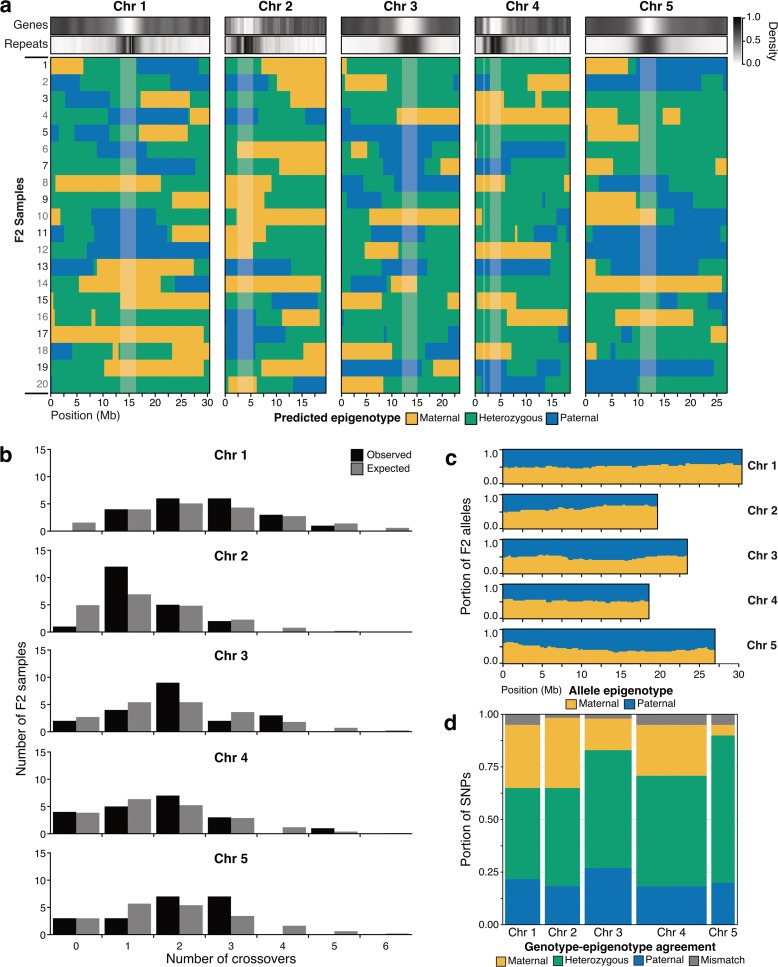
Fig. 4.
Results of epigenotyping in F2 samples. a Final epigenotype maps for all chromosomes in the F2 population of A. thaliana Col-0 MA line 49 × line 69. Yellow indicates homozygous maternal, green heterozygous, and blue homozygous paternal. Lighter color background denotes the centromere. Heatmaps above chromosomes show gene and repeat density across the chromosome. b Distribution of observed number of crossovers per chromosome, in black, compared to expected number based on Poisson distribution, in gray. c Epigenotype predicted allele frequencies across each chromosome. From the law of segregation, maternal (yellow) and paternal (blue) alleles are expected in a 1:1 ratio. d Agreement of predicted epigenotype based on epigenotype map and predicted genotype based on reads for F2 samples at SNPs. Width of bar indicates number of SNPs on the chromosome. Yellow, green, and blue indicate both methods predicted maternal homozygous, heterozygous, and paternal homozygous, respectively. Gray indicates mismatch between the methods