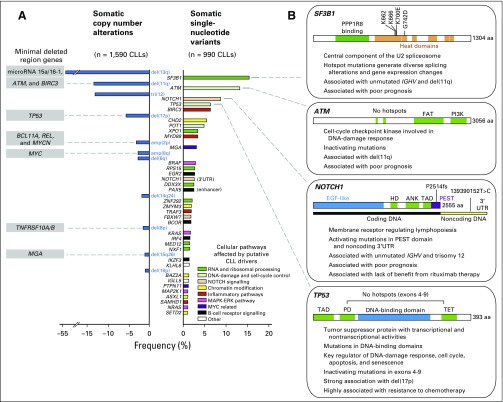
Fig 1.
Putative driver gene mutations and recurrent somatic copy number variations in chronic lymphocytic leukemia (CLL). (A) Frequency of somatic copy number alterations when combining cohorts from the Spanish International Cancer Genome Consortium (ICGC) trial12 (n = 452), German CLL Study Group CLL8 trial11 (n = 353), Scandinavian SCALE trial13 (n = 369), Ouillette et al10 study (n = 255), and Dana-Farber Cancer Institute (DFCI)/Brown et al9 study (n = 161), identified through single-nucleotide polymorphism array (except as identified by whole-exome sequencing [WES] for Spanish ICGC cohort). Also shown are the frequencies of somatic single-nucleotide variants (from DFCI/Broad Institute14 [n = 548] and from Spanish ICGC12 [n = 452]) identified through WES. Only the events over 1% in frequency in the combined cohorts are reported. IGLL5, MAP2K1, and SAMHD1 have been reported in DFCI cohort only, whereas ZNF292, KLH6, SETD2, and PAX5 enhancer have been reported in Spanish ICGC cohort only. All remaining genes were consistent among cohorts. Mutations in the 3′UTR of NOTCH1 were detected in four of 150 patient cases with whole-genome sequencing. (B) Summaries of the characteristics of frequently mutated genes in CLL. amp, amplification; ANK, ankyrin repeat; del, deletion; EGF, epidermal growth factor; ERK, extracellular signal-regulated kinase; FAT, FRAP-ATM-transformation/transcription domain associated protein; HD, heterodimerization domain; MAPK, mitogen-activated protein kinase; PD, programmed death; PEST, proline, glutamic acid, serine, and threonine; PI3K, phosphatidylinositol 3-kinase; SCALE, Scandinavian Lymphoma Etiology; TAD, transaction activation domain; TET, ten-eleven translocation.