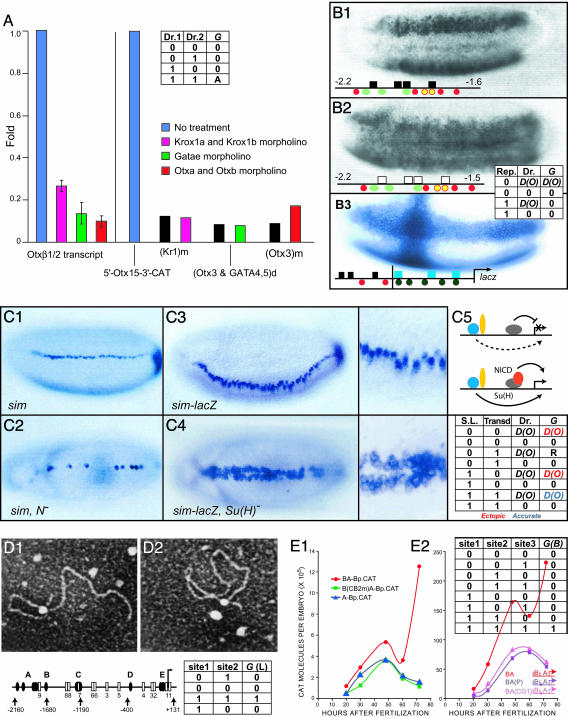
Fig. 1.
Cis-regulatory logic functions. (A) Experimental example of AND logic operation. [Reproduced with permission from ref. 14 (Copyright 2004, Elsevier).] Bars display relative activity of expression constructs, including a cis-regulatory element of the sea urchin otx gene, driving a chloramphenicol acetyltransferase (CAT) reporter. The construct was injected into sea urchin eggs, and the level of CAT transcripts it produced was measured by using quantitative PCR. Results of removal of individual inputs by antisense (morpholino) treatment are shown for wild-type construct as indicated (colored bars), or, when individual respective target sites were mutated (black bars). As required, blocking the inputs and mutating the sites gives the same effect; all three inputs (Kr, Gata, and Otx) are needed, and if any is absent, no significant activity is obtained (14). The truth table shows values of the G operator function for a two input system [drivers 1 and 2 (Dr. 1 and Dr. 2)], where 0 and 1 indicate sub- and above-threshold inputs respectively, and A indicates an activating output (see text). (B) Experimental example of short-range repressor operation. Photographs display lacZ reporter staining in transgenic Drosophila embryos bearing rhomboid (rho) expression constructs. Relevant target sites are indicated below. Activators: Dorsal (red), bHLH (green), Twist (yellow), and Bicoid (black circles). Repressors: Snail (black boxes) and Krüppel (blue boxes). (B1 and B2) Ventral views. Expression of rho-lacZ constructs with and without target sites (open boxes, mutated) for the Snail short-range repressor is shown. These sites are required to prevent expression in the prospective mesoderm. [Images in B1 and B2 are reprinted with permission from ref. 30 (Copyright 1994, Cold Spring Harbor Lab. Press).] (B3) Autonomy of short-range repression, demonstrated by fusion of rho and evenskipped stripe 2 (eve2) CRMs; the latter uses the Krüppel short-range repressor to establish posterior border of expression. The crossed expression pattern shows that each repressor functions independently, and that neither repressor interferes with the activation of the other CRM. [The image in B3 is reprinted with permission fromref. 15 (Copyright 1996, Cold Spring Harbor Lab. Press).] The truth table shows that the operator function G has the activating regulatory value produced by the level of driver occupancy [D(Oc(s, t))], here for a one-driver element}, whereas if the repressor (Rep) is present, or the driver is absent, there is no output. (C) Transcriptional toggle switch, Notch (N) signal transduction system effects on the single-minded (sim) gene of Drosophila (data are from ref. 31). (C1) Wild-type endogenous expression of sim in prospective midline neuroblasts. (C2) Expression of sim in N mutant embryos; N signaling is required positively for normal expression.(C3) Expression of sim-lacZ construct in wild-type embryos; an enlargement is on the on right. (C4) A view of C3, in embryos lacking the transcription factor [Su(H)] that transduces the N signal. In the absence of Su(H), ectopic expression occurs. (C5) CRM diagrams: Dorsal and Twist, activators (blue and yellow, respectively), Su(H), a repressor (gray), except when bound by intracellular N fragment (red), as a result of N signaling. [Images in C1–C5 are reprinted with permission from ref. 31 (Copyright 2000, Cold Spring Harbor Lab. Press).] The truth table shows that there are several possible values of the operator function G: expression, ectopic, or normal (according to the level of driver occupancy, here treating the two molecules portrayed as a single driver), or repression of output that would otherwise be produced according to D(O) (R, only in those cells where the values 0, 1, [D(O) obtain], or just no output (0). The values are combinatorially conditional, depending on whether the signal ligand (S.L.) is presented, and whether or not the transducer [here, as Su(H)], and the activating driver (e.g., here, Twist or Dorsal) are present. (D) Loop formation mediated by multiple CRM sites (17). (D1) Map of cis-regulatory system of the sea urchin cyIIIa gene, with specific sites for SpGCF1, a multimerizing, DNA-binding protein is highlighted; sites for other transcription factors are shown as as open boxes. (D2 and D3) Electron micrographs of loops formed by purified SpGCF1 protein mixed with cyIIIa cis-regulatory DNA in vitro.(D2) A–C site loop. (D3) A–E site loop. [Images in D1–D3 are reprinted with permission from ref. 17 (Copyright 1995, National Academy of Sciences).] The truth table illustrates the point that loop formation, an all-or-nothing operation, requires both sites occupied for any given loop to form. (E) Combinatorial functional linkage of two CRMs, data are from the sea urchin endo16 gene (3, 9). Kinetics for output of CAT reporter enzyme when module A (proximal) and the adjacent module B are included in the construct, are shown in red over developmental time, in two experiments. (E1) Kinetics for module A alone (blue), are identical with kinetic output of whole BA construct bearing mutation of site for a CRM B-CRM A DNA-binding linker protein (CB2). [The image in E1 is reprinted with permission from ref. 9 (Copyright 1998, AAAS, www.sciencemag.org).] (E2) Mutations of sites for two other linker proteins (P and CG1) in the otherwise complete BA construct yield kinetics that are also the same as for CRM A alone. [The image in E2 is reprinted with permission from ref. 3 (Copyright 2001, Company of Biologists Limited).] The truth table shows that all three sites are required for normal kinetic input of module B [here, abbreviated as G(B)] to register, and only in this condition are the red curves generated.