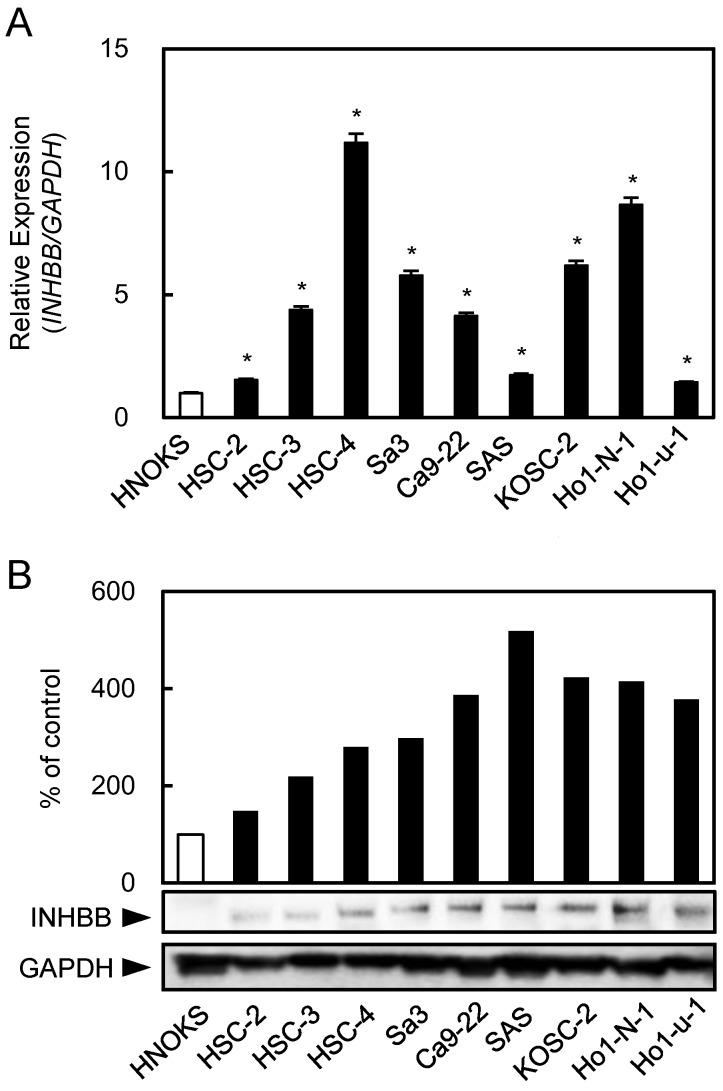
Figure 1.
Up-regulation of INHBB expression in OSCC-derived cell lines. (A) Quantitation of INHBB mRNA expression by qRT-PCR analysis in OSCC-derived cell lines. Significant (*P < 0.05, Student's t-test) up-regulation of INHBB mRNA is observed in nine OSCC-derived cell lines compared to the HNOKs. The data are expressed as the mean ± SEM of triplicate results. (B) Western blotting of INHBB protein in OSCC-derived cell lines and HNOKs. INHBB protein expression is up-regulated in OSCC-derived cell lines compared with that in the HNOKs. Densitometric INHBB protein data are normalized to the GAPDH protein levels. The values are expressed as a percentage of the HNOKs.