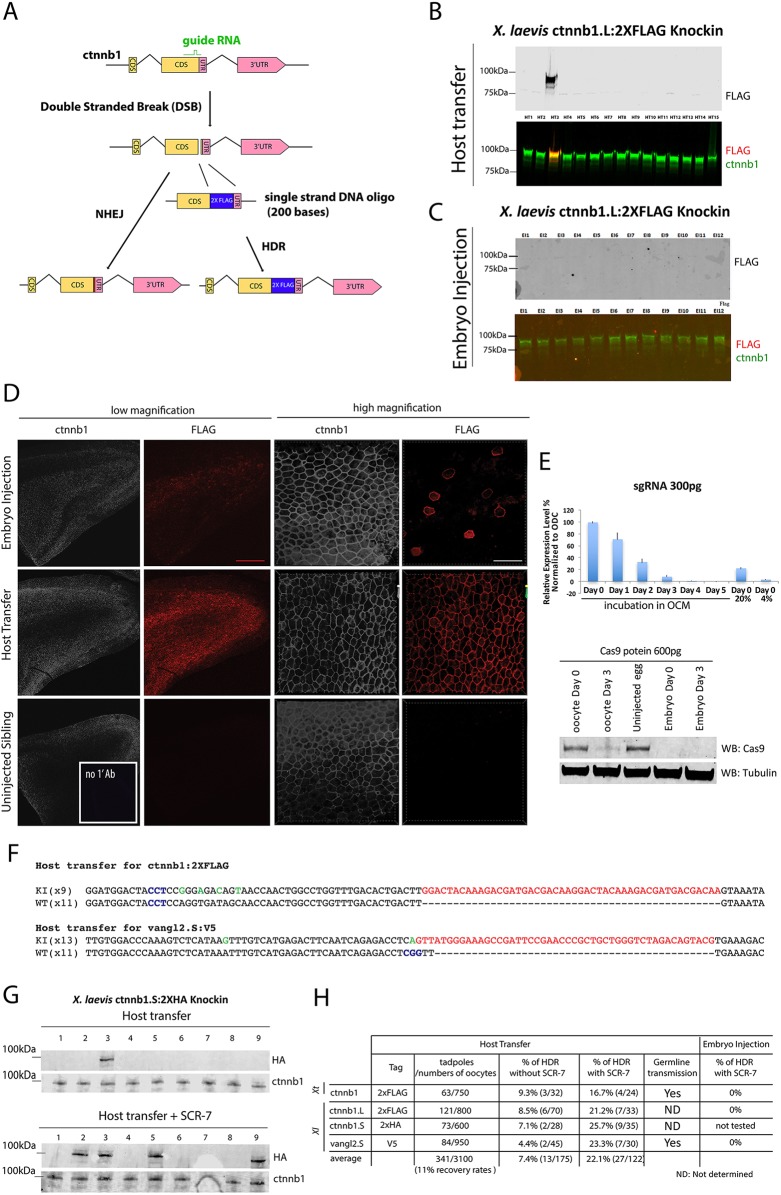
Fig. 2.
Comparative analysis of HDR in Xenopus oocytes and embryos. (A) The procedure for precisely inserting a 2×FLAG tag into the ctnnb1 gene through an HDR-mediated recombination event. (B) Representative western blot of single X. laevis embryos obtained by oocyte injection/host transfer. In the bottom image the green bands represent β-catenin protein and the red band represents the FLAG tag. (C) Representative western blot with of X. laevis embryos derived by embryo injection. (D) Whole-mount immunostaining of epithelial tissues shows expression of the FLAG tag in the same pattern as β-catenin protein in oocyte-injected samples, as expected for heterozygous embryos, and in just a few cells in embryo injected samples, as expected for mosaic embryos. Scale bars: 250 μm (red); 30 μm (white). (E) Real-time PCR (top) shows the stability (mean±s.d. from three independent experiments) of sgRNA in oocytes and western blot (bottom) demonstrates the decay of 600 pg Cas9 protein injected into oocytes and embryos of X. laevis. (F) Sequencing of 20 (for 2×FLAG) or 24 (for V5) clones from the oocyte-injected embryo shows successful knock-in of 2×FLAG in one allele of ctnnb1 and of the V5 epitope in one allele of vangl2.S. PAM sequences are marked in blue; green indicates mismatched nucleotide from the RO; red indicates epitope. No conclusions were drawn from lane 7 owing to damage to the blot. (G) Treatment of oocytes with the DNA ligase IV inhibitor SCR-7 (5 µM) increases the efficiency of successful HDR-mediated knock-in. Representative western blots are shown for ctnnb1.S:2×HA with or without SCR-7 treatment. (H) The overall efficiency of HDR events in F0 tadpoles. Xt, X. tropicalis; Xl, X. laevis.