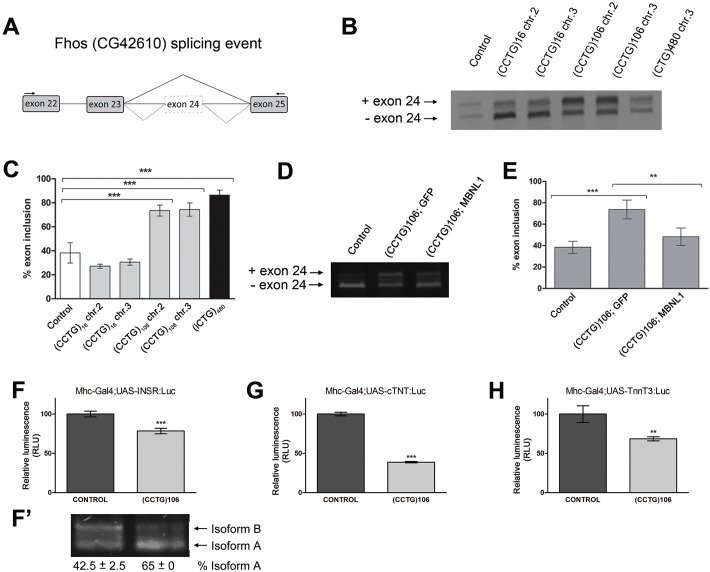
Fig. 2.
DM2-106 expression in muscle causes mis-splicing of MBNL1-dependent transcripts. (A) Outline of the intron/exon structure of Fhos (CG42610) showing the exons implicated in the splicing event studied. Wild-type flies mainly skipped exon 24 (solid line), whereas DM2-106 expression in IFM led to aberrant inclusion of exon 24 (dotted lines). Arrows indicate primers used for semi-quantitative PCR analysis. (B,C) Agarose gel and quantification of Fhos RT-PCR products from IFM expressing control (N-16) and DM2-106 transgenes located on chromosomes 2 and 3. These transgenes were driven by Mhc-Gal4. Flies that only contain the Mhc-Gal4 driver without a UAS transgene show an average frequency of exon 24 inclusion of ∼30%. Compared with this control, expression of normal repeat length (CCUG)16 does not significantly alter Fhos splicing, whereas in the (CCUG)106 repeat-expressing cells exon 24 is retained at ∼70%, levels similar to those of DM1 flies expressing an interrupted 480 CUG repeat sequence (iCUG)480. (D,E) Agarose gel and quantification of Fhos RT-PCR products from flies expressing the indicated transgenes with the Mhc-Gal4 driver. Simultaneous expression of human MBNL1 and DM2-106 induces exon 24 exclusion, restoring wild-type levels (Mhc-Gal4 only). Error bars represent s.d. and each experiment was repeated at least twice in adults of 0-5 days of age. (F-H) Luminescence levels of Mhc-Gal4>UAS-minigene,DM2-106 normalized to the levels of Mhc-Gal4>UAS-minigene,UAS-GFP. Relative luminescence decreased from 100% in control flies to 78% for the human INSR reporter minigene (F), 38% for TNNT2 (cTNT) (G) and 68% for mouse Tnnt3 (H). RLU, relative light units. **P<0.005, ***P<0.001 (Student's t-test). (F′) RT-PCR analysis of the INSR spliceosensor in N-16 and DM2-106 background. The percentage is the average of two experiments.