Figure 4. Tiny amounts of data exert decisive influence in the resolution of certain contentious branches in phylogenomic studies.
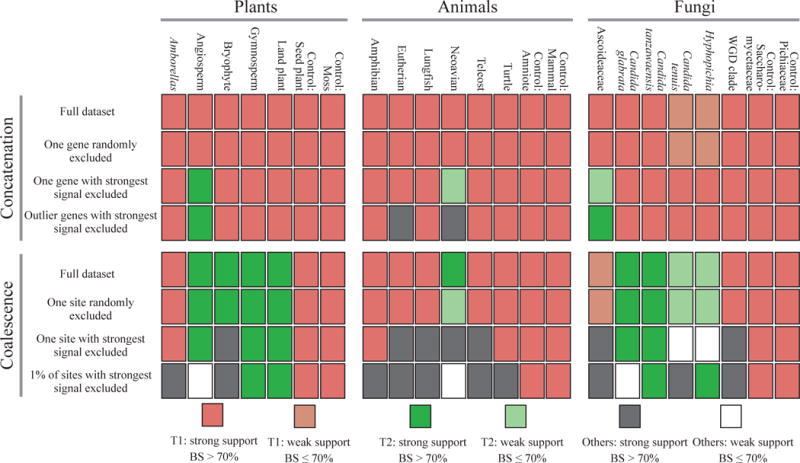
The effect of the removal of tiny amounts of data on the branch’s topology and bootstrap support (BS) was quantified for 17 contentious branches and 6 well-established branches (controls) in plant, animal, and fungal phylogenomic data matrices. Different colors indicate different branch topologies and levels of BS. Topologies other than T1 and T2 are collectively referred to as “Others”. (Top panel: Concatenation) The first row depicts the results of the concatenation analysis when the full data matrix is used, the second row when a single random gene is excluded, the third row when the gene with the highest absolute ΔGLS value is excluded, and the fourth row when the genes with outlier ΔGLS values are excluded. (Bottom panel: Coalescence) The first row depicts the results of the coalescence-based analysis when the full data matrix is used, the second row when one random site from every gene’s alignment is excluded, the third row when the site with the highest absolute ΔSLS value from every gene is excluded, and the fourth row when the 1% of sites with the highest absolute ΔSLS values from every gene are excluded. All topologies summarized in this figure are provided in Supplementary Figs. 5–56.
