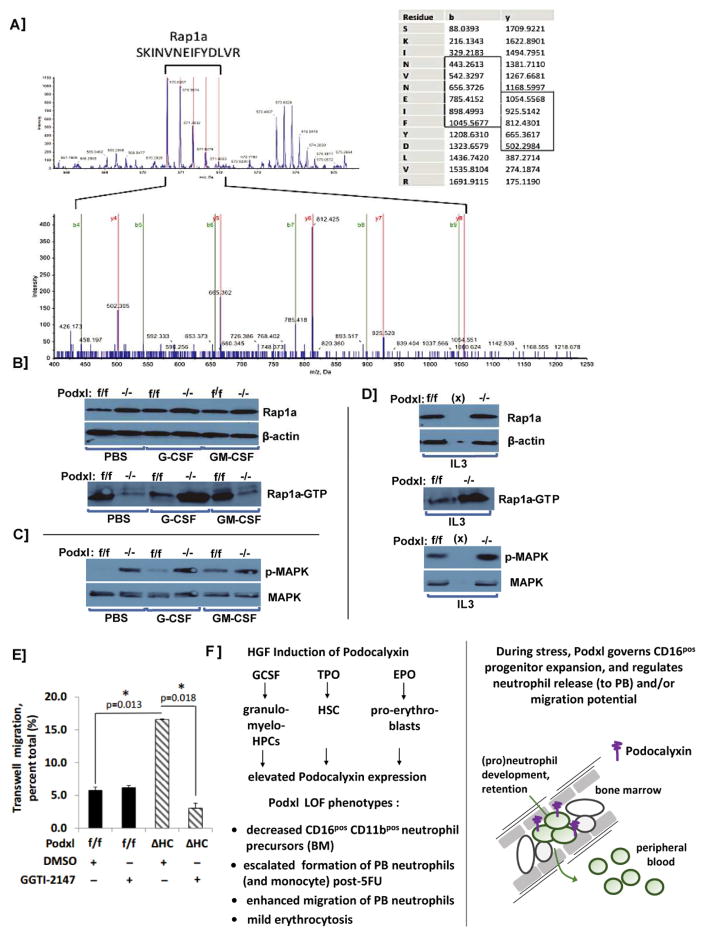
Figure 3. Podxl interacts with Rap-1A in HPCs, and regulates Rap1aGTP levels.
A] Following G-CSF dosing of wild-type mice, hematopoietic progenitor cells (HPCs) were isolated from bone marrow cell preparations as Linneg populations (depleted for CD5, CD11b, CD19, CD45R, Ly6G/C and Ter119). From cell lysates, Podxl (co-)immunoprecipitates were then prepared, reduced, alkylated and used to generate tryptic peptides. Peptides were analyzed by LC-MS/MS. For one predominant Podxl partner, Rap1a, representative time of flight mass spectra (MS) are shown (e.g., SKINVNEIFYDLVR). The left panel identifies precursor peptide ion mass 1708.9148, z=3 and delta mass= −0.0001 (99% confidence). The right panel summarizes collision-induced decay fragment spectra for sequence determination of SKINVNEIFYDLVR. B] Podxl regulates levels of activated Rap1a-GTP: Hematopoietic progenitor cells were prepared from the bone marrow of Podxlf/f and PodxlΔHC mice, and plated in IMDM (1x106cells/mL). Here, Linneg bone marrow HPCs from wild type and Podxl-KO mice were used. Cells were then exposed to PBS, G-CSF, or GM-CSF. At 30 minutes, lysates were generated and analyzed by western blotting for total Rap-1A levels, and levels of GTP-bound Rap-1A (lower panel). C] For the above samples (4B), levels of p-MAPK and total MAPK also were assessed. D] Podxl-KO dysregulates IL3- induced Rap1aGTP formation: HPCs isolated from Podxl+/+ and PodxlΔHC mouse bone marrow. Following HGF withdrawal, cells were stimulated with IL3 (10ng/mL, 30 minutes). Cell lysates were then prepared, and levels of Rap1a and Rap1aGTP were determined (as in 4B above). E] Podxl deletion enhances neutrophil migration: Peripheral blood neutrophils were prepared via density gradient centrifugation using Histopaque-1077 and Histopaque-1119. Trans-well migration of peripheral blood neutrophils from wild type and Podxl-KO mice was then assessed +/− the Rap1a inhibitor GGTI-2147. Graphed results are mean values +/− SE (n=3) for trans-membrane migrating cells. F] Initial model for Podxl governing of stress myelopoiesis: Left panel: HGF effects on Podxl expression are outlined (upper sub-panel), together with Vav1-conditional Podxl-KO phenotypes. Right panel: A working model is outlined at a cellular level for proposed Podocalyxin governing of peripheral blood neutrophil (e)migrations.