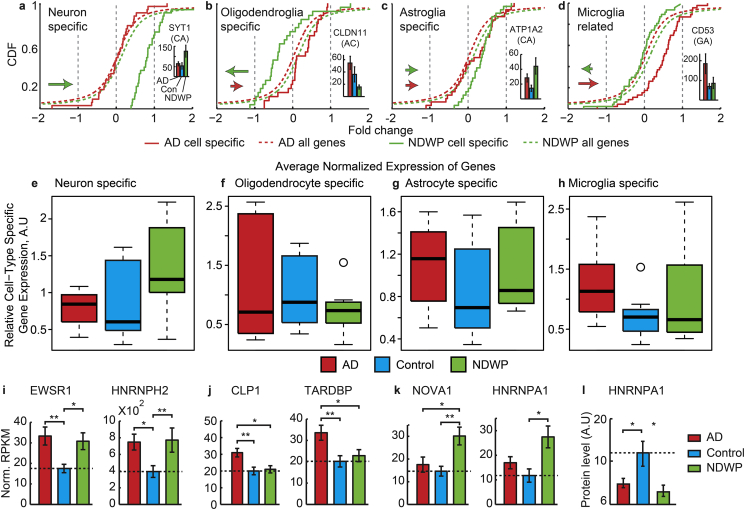
Fig. 4.
In-depth sequencing identifies cell type-specific, cognition- and pathology-associated transcript differences. (a–h) NDWP NBB brains show up-regulated neuronal transcripts and down-regulated transcripts in other cell types. (a–d) Shown are cumulative distribution functions (CDFs) for the global change between AD or NDWP and control (dashed lines, red and blue, correspondingly) for the subgroups of cell type-specific genes, following Cahoy et al. (Cahoy et al., 2008) (solid lines, same colors). (a) Neuronal genes are sharply up-regulated in NDWP (a; Kolmogorov Smirnov (KS) P value < 0.0001). Columns: APA variant of the calcium sensor SYT1 with a 3′-CA terminus. (b) Oligodendroglia genes were down regulated in NDWP and up-regulated in AD (KS P value < 0.001). Columns: a 3′-AC-terminated variant of the central nervous system myelin-associated CLDN11 gene. (c) Astroglial genes were moderately up-regulated in both disease groups (KS P value < 0.01). Columns: a CA variant of the metabolic regulator ATP1A2. (d) Microglia-related genes were up regulated in both NDWP and AD. Columns: a 3′- GA variant of the differentiation marker CD53 and following Zhang (Zhang et al., 2013). (e–h) Parallel box plot presentation of normalized values for a different list of human cell-type characteristic genes, following Darmanis (Darmanis et al., 2015) with healthy control levels referred to as 1. Note parallel differences in this analysis as well. (i) AD-related differences in RNA metabolism genes: normalized RPKM values across the three patient groups (mean ± SEM) for EWSR1 and HNRNPH2 (One-way-ANOVA P values = 0.02 and 0.04, correspondingly), CLP1 and TARDBP (One-way-ANOVA P values = 0.005 and 0.02, correspondingly), and NOVA1 and HNRNPA1 (One-way-ANOVA P values = 0.005 and 0.02, correspondingly).