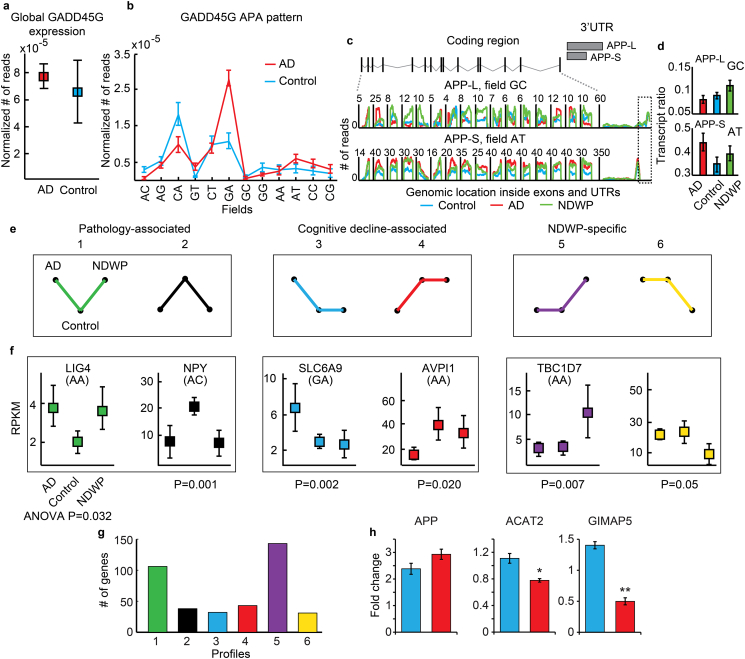
Fig. 6.
APA variants classify patient groups better than cumulative transcripts from all fields together. (a) Expression levels of GADD45G in AD patients and controls. (b) APA pattern of GADD45G in AD (red) and controls (blue). Shown are averages ± SEM of expression in each SQUARE field in the patient groups, note switch between the proximal and distal products from controls to AD. Fields ‘CA’ and ‘GA’, but none of the others neither the global transcript counts show significant change. (c) APP gene structure with two APA variants; one with a long (APP-L) and one with a short 3′-UTR (APP-S). Numbers of sequencing reads across APP exons in SQUARE fields ‘GC’ and ‘AT’. The unique 3′-UTR region of APP-L is marked in a dashed rectangle. (d) Number of reads in specific SQUARE fields divided by the number of total transcript reads in each sample for APP-L and APP-S. In both cases One-way-Anova-P value has been < 0.05. (e) Predicted profiles of expression differences between the groups may reflect association with pathology, cognitive decline or NDWP-specific features. (f) Mean ± 95% confidence level for the exemplary genes LIG4, NPY, SLC6A9, AVPI1, TBC1D7, and NDUFA3, each presenting a distinct profile (colors as in a). (g) Bar graph for transcript APA counts in each profile (colors as in e). (h): Normalized luciferase activity for transcripts harboring the long (L – blue bars) or the short (S – red bars) 3′UTRs of APP, ACAT2 and GIMAP5. Briefly, the complete 3′-UTRs of human APP, ACAT2 and GIMAP5 were amplified from genomic DNA, and cloned downstream of the Renilla luciferase gene in psiCHECK2 (Clonetech) using XhoI and NotI. To create vectors expressing the short 3′-UTR alone, this region of the 3′- UTR from each of the cloned transcripts was similarly sub-cloned into psiCHECK2. To obtain expression of the long 3′ UTR isoform alone, the proximal polyadenylation site was mutated from AAUAAA to ACUCAA (APP, ACAT2) and AAUAGA to AAUCGA (GImAP5) using QuickChange Site Directed Mutagenesis (Agilent). HEK-293 cells were cultured in DMEM supplemented with 10% fetal bovine serum and transfected with TransIT-X2 (Mirus) according to the manufacturer's instructions. Cells were then incubated overnight before performing luciferase assays. Luciferase activity was assessed using the Dual-Glo system (Promega) performed according to the manufacturer's instructions. Renilla fluorescence was normalized to firefly signal, and results are presented as this ratio.