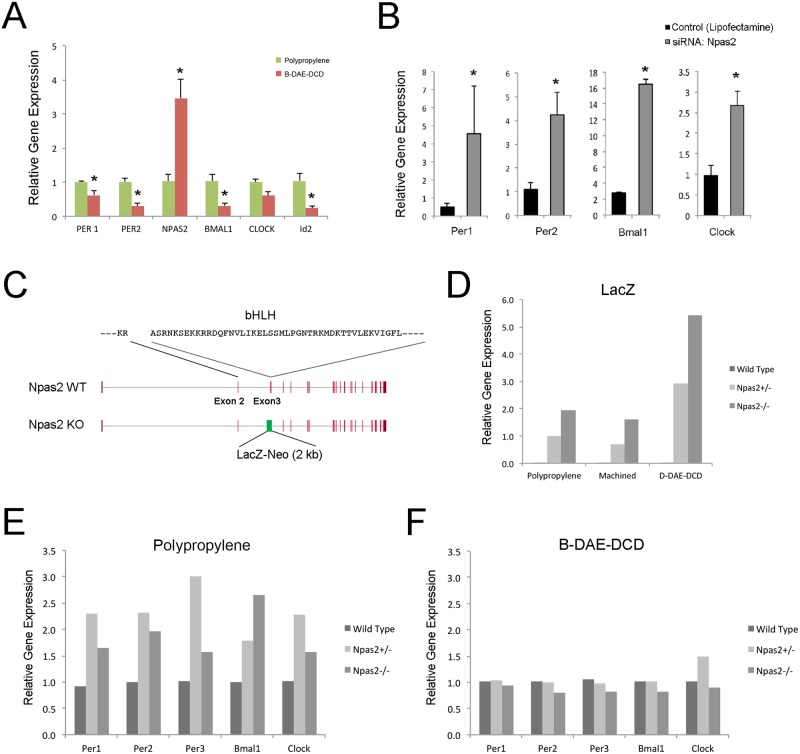
Fig 4. The role of Npas2 in BMSC circadian rhythm.
A. A snapshot of circadian rhythm gene expression from rat BMSCs shows the transcriptional downregulation of Per1, Per2, Bmal1 and Id2 when the BMSCs were cultured on B-DAE-DCD Ti discs. A similar upregulation in Npas2 was observed in human BMSCs. RT-PCR was performed in triplicates. *: p<0.05 by Student’s t-test. B. The role of the transcription factor Npas2 on the steady state mRNA levels of circadian rhythm-related genes was examined by siRNA. Npas2 knock down appeared to have a greater impact on circadian rhythm-related gene expression in BMSCs cultured on the polypropylene dishes. *: p<0.05 by Student’s t-test. C. To further clarify the role of Npas2, BMSCs were harvested from mice carrying the Npas2 allele lacking Exon 3, which was replaced by the LacZ reporter gene cassette, resulting in a Npas2 functional knockout mutation due to the lack of the basic helical-loop-helical (bHLH) domain. D. BMSCs were harvested from the femurs of wild-type, Npas2+/- and Npas2-/- male mice (n = 5 each). BMSCs (passage 4) were cultured and synchronized on polypropylene dishes, machined Ti discs or B-DAE-DCD discs (n = 2 in each group). Thirty-two hours after synchronization, LacZ reporter gene expression was increased in the D-DAE-DCD disc group. E. The expression of circadian rhythm genes were observed to increase in the Npas2+/- and Npas2-/- BMSCs maintained on polypropylene culture dishes for 32 hours after synchronization. F. Npas2+/- and Npas2-/- BMSC maintained on B-DAE-DCD discs for the same length of time demonstrated similar circadian rhythm gene expression patterns as wild-type mouse BMSCs.