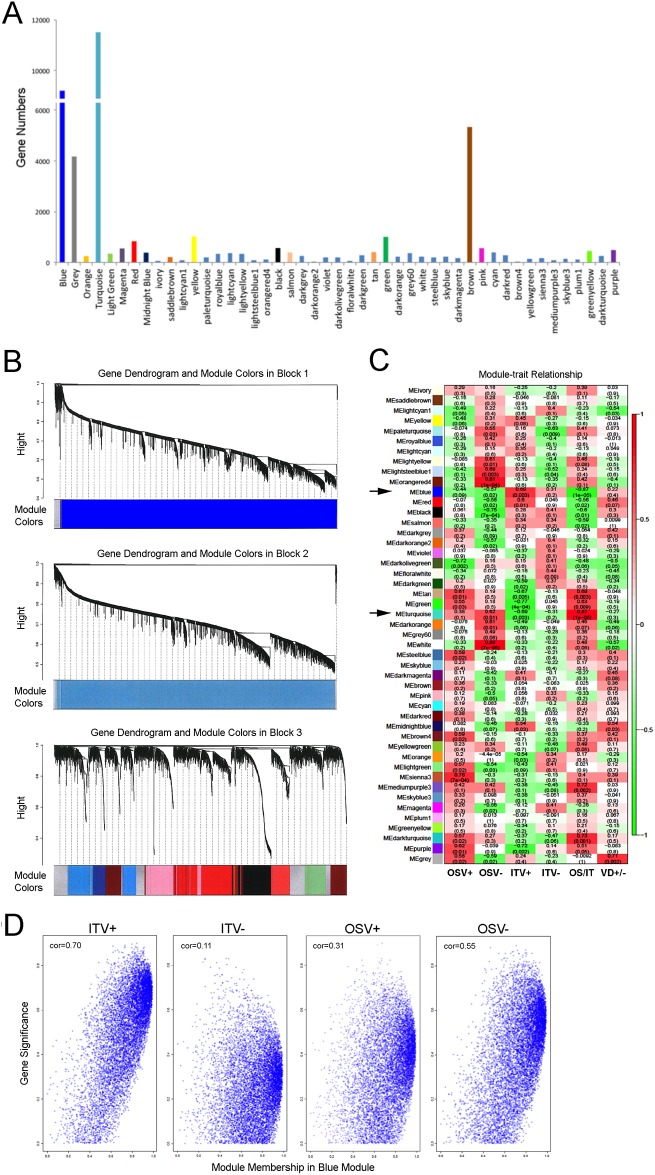
Fig 5. The weighted gene co-expression network analysis (WGCNA).
The microarray data were obtained previously from rat femur bone marrow tissue after osteotomy surgery with or without DAE-DCD Ti implants with sufficient or deficient serum vitamin D levels (ITV+, ITV-, OSV+ and OSV- groups, respectively) [20]. A. WGCNA identified 47 modules of co-expressed genes. The Turquoise and Blue modules contained a disproportionately large number of genes. B. A dendrogram of the WGCNA analysis suggested an unusually large co-expressed gene network of hierarchical clustering among Blue and Turquoise modules, which contained 9,202 and 11,511 genes, respectively. C. Association of the module eigengenes with experimental bone marrow tissue traits responding to osteotomy surgery with or without Ti implant placement (IT and OS, respectively) in vitamin D-sufficient and -deficient rats (V+ and V-, respectively). Out of the 47 modules identified, the Blue and Turquoise modules (arrows) showed the highest eigengene correlation in OS/IT. Within each cell, upper values indicate correlation coefficients between the module eigengene and the traits, while the lower values indicate the corresponding p-value. D. Scatter plots of module membership (eigengene-based connectivity) and gene significance for the Blue module for each of the therapeutic traits (ITV+, ITV-, OSV+ and OSV-). The highest correlation between module membership and gene significance was in ITV+; this was noticeably decreased in ITV-.