Abstract
Honey bees are important pollinators of agricultural crops. Pathogens and other factors have been implicated in high annual losses of honey bee colonies in North America and some European countries. To further investigate the relationship between multiple factors, including pathogen prevalence and abundance and colony health, we monitored commercially managed migratory honey bee colonies involved in California almond pollination in 2014. At each sampling event, honey bee colony health was assessed, using colony population size as a proxy for health, and the prevalence and abundance of seven honey bee pathogens was evaluated using PCR and quantitative PCR, respectively. In this sample cohort, pathogen prevalence and abundance did not correlate with colony health, but did correlate with the date of sampling. In general, pathogen prevalence (i.e., the number of specific pathogens harbored within a colony) was lower early in the year (January—March) and was greater in the summer, with peak prevalence occurring in June. Pathogen abundance in individual honey bee colonies varied throughout the year and was strongly associated with the sampling date, and was influenced by beekeeping operation, colony health, and mite infestation level. Together, data from this and other observational cohort studies that monitor individual honey bee colonies and precisely account for sampling date (i.e., day of year) will lead to a better understanding of the influence of pathogens on colony mortality and the effects of other factors on these associations.
Introduction
Honey bees (Apis mellifera) are the primary insect pollinators of agricultural crops, including fruits, nuts, and vegetables, which have an approximate annual value of $17–18 billion in the United States [1,2]. California almond production is the most renowned example of the role of honey bee pollination services in the US. Every year, over 60% of the approximately 2.5 million commercially managed honey bee colonies in the United States are transported to the Central Valley of California to pollinate the almond crop. California almond growers produce ~ 80% of the global almond supply, which was valued at $1.7 billion in 2014 [3]. Honey bee colonies are important for the pollination of plants in both agricultural and non-agricultural landscapes [4,5]. Thus, high annual honey bee colony losses in the US, which have averaged ~ 33% annually since 2006 and increased from historic levels of approximately 12%, are concerning to stakeholders, including beekeepers, growers, and scientists [6,7]. To compensate for increased colony losses and meet pollination service demands, beekeeping operations have been splitting their colonies (i.e., producing two colonies from one colony) more frequently. Therefore, the total number of commercially managed honey bee colonies in the US has been maintained at approximately 2.5 million since the late 1990s [8] despite unsustainable annual colony losses.
There are multiple factors that impact colony health, including pathogens (i.e., mites, viruses, bacteria, and fungi), queen failure, colony genetics, weather, nutrition, management practices (i.e., transportation, treatments), and agrochemical exposure [7,9–18]. Honey bee colony health is typically evaluated by estimating colony population size [19–23]. Colonies are comprised of ~35,000 sterile female workers, hundreds of males (drones), and a single reproductive queen that can lay approximately 1,000 eggs per day [24]. While no single factor has been deemed responsible for high annual honey bee colony losses, pathogens are major contributing factors [6,7,10,11,21,22,25–33].
Honey bees host a diversity of pathogens, the majority of which are single-stranded RNA viruses [33,34]. Honey bee infecting viruses include Acute bee paralysis virus (ABPV), Black queen cell virus (BQCV), Deformed wing virus (DWV), Israeli acute paralysis virus (IAPV), Kashmir bee virus (KBV), Sacbrood virus (SBV), Chronic bee paralysis virus (CBPV) [34–38] and the Lake Sinai viruses [23]. In addition to viruses, pathogens of honey bees include eukaryotes such as the trypanosomatid Lotmaria passim (formerly Crithidia mellificae strain sf [39,40]), the microsporidial pathogen Nosema ceranae [41], and bacterial pathogens such as Paenibacillus larvae [42] and Melissococcus plutonius [43], the causative agents of American and European foulbrood diseases, respectively. In addition, the ectoparasitic mite, Varroa destructor, contributes to decreased colony health by feeding on developing bees (brood) and facilitating virus transmission [44] (i.e., DWV [14,45], KBV [46,47], and IAPV [34,44,48]). Mite parasitization of developing honey bees can result in physical deformities, reduced body weight, greater DWV virus levels, and/or death [44,49].
Temporal monitoring studies are essential for identifying and understanding the factors that influence honey bee colony health and pathogen prevalence and abundance. Identifying particular pathogens or suites of pathogens that are associated with colony losses is complicated because pathogen prevalence and abundance varies by season and geographic location [21,22,25,26,31]. Furthermore, there have been relatively few studies that have monitored individual commercially managed colonies [7,10,15–17,21,22]. The majority of studies in the US have monitored honey bee health at the apiary level with the main goals of detecting exotic pathogens and establishing a baseline understanding of honey bee colony health [6,7,15,17,30]. Additional studies that monitor colonies located in varying geographic locations are needed to better understand the impacts of multiple factors on honey bee colony health. To address this knowledge gap, we monitored Minnesota-based commercially managed honey bee colonies involved in the 2014 California almond pollination event. The goal of this study was to investigate the association between multiple factors, including pathogen seasonality, pathogen abundance, beekeeping operation, colony population size, and level of mite infestation, on honey bee colony health and colony losses. We determined that sampling date strongly influenced pathogen prevalence and abundance and confirmed a strong association between DWV abundance and Varroa destructor mite infestation. However, the abundance of other viruses (i.e., BQCV, LSV1, and LSV2) was not associated with mite infestation. Together, data from this and other temporal observational cohort studies that precisely account for sampling date (i.e., day of the year) and monitor individual honey bee colonies will lead to a better understanding of the association between factors affecting pathogen abundance and honey bee colony health.
Methods
Longitudinal monitoring and sampling of commercially managed honey bee colonies
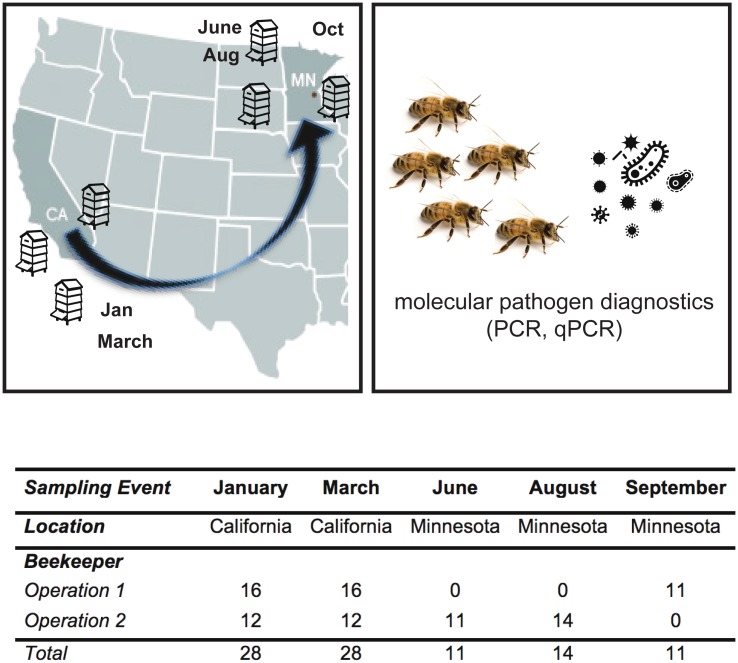
Honey bee (Apis mellifera) colonies from two Minnesota-based commercial beekeeping operations that transport honey bee colonies to California for almond pollination were monitored for one year (i.e., from January 2014 to January 2015). At the onset of the study in January 2014, beekeepers identified 20 colonies that successfully overwintered. Samples from a subset of these colonies were analyzed for pathogen prevalence and abundance (n = 28), of which two died before November 2014, and were replaced by three new colonies bringing the total unique colonies described in this dataset to 31. Each beekeeping operation administered anti-mite treatments. To reduce Varroa destructor mite infestation levels in 2014, Operation 1 treated colonies with amitraz on April 22 and Sept. 4, and oxalic acid on Oct. 20, and Operation 2 treated colonies with amitraz on March 15, oxalic acid on Aug. 21, amitraz on Sept. 9, and formic acid on Oct. 24. Colony health, using colony population size as a proxy for health, was estimated by counting and imaging the number of frames more than two-thirds covered with bees. Colony health was categorized as weak (< 7 frames), average (7–12 frames), or strong (> 12 frames) based on frame count (S3 Table) [20,21]. Samples of honey bees were obtained from monitored colonies, up to four times each. Specific 2014 sample dates varied and were categorized into discrete sampling events in order to better compare data within and between colonies managed by two beekeeping operations. Specifically, “before almond pollination” samples were obtained from colonies located in California in January 2014 (Beekeeping Operation 1 on Jan. 27 and Beekeeping Operation 2 on Jan. 29). Samples that represent the pathogens associated with honey bee colonies “during almond pollination” were obtained immediately post-almond pollination in March 2014 (Beekeeping Operation 1 on March 3 and Beekeeping Operation 2 on March 14). “After almond pollination” samples were obtained from colonies that were located in Minnesota in early (June 9) and late (Aug. 25 and Sept. 2) summer (Fig 1 and S3 Table). Additionally, core samples containing pollen/bee bread, wax, and honey were obtained from a representative frame in each colony and analyzed for pesticide residues, as described in Kegley et al. 2017 [50]. The sample cohort analyzed herein includes pathogen prevalence, abundance, and survivorship data obtained in 2014, including variable numbers of samples obtained from dead (n = 2), weak (n = 8), average (n = 34), and strong (n = 48) rated colonies (total = 92) spanning the duration of the 2014 California almond pollination season, and colony longevity data through January 2015 (Fig 1 and S3 Table). In total, 21 colonies in this sample cohort died by the end of January 2015.
Fig 1. Commercially managed honey bee colonies were longitudinally monitored before, during, and after the 2014 almond pollination season.
Honey bee colonies from two Minnesota-based commercial beekeeping operations that transport their colonies to California for almond pollination were monitored from January 2014 to January 2015. At each sampling event, colony health, using colony population size as a proxy for health, was monitored and samples of live honey bees were obtained. In a subset of samples described in the table, PCR was utilized to assess pathogen prevalence and qPCR was utilized to determine pathogen abundance.
Honey bee samples
At each sampling event, live honey bees (~ 150 per sample) were obtained from a frame containing developing bees (brood) in the center each colony. Samples were composed of female bees of mixed age. Collected samples were placed on ice or dry ice in the field, stored at -20°C, transported on dry ice, and transferred to -80°C for storage prior to analysis. Five female bees from each sample were used for RNA extraction, cDNA synthesis, pathogen-specific PCR, and qPCR. The objective for pathogen screening was to identify the most prevalent pathogens associated with honey bees sampled from individual colonies at each sampling event. Based on empirical data, literature values, and practical sample handling considerations, we assayed five bees per colony per sampling event [22]. The following equation from Pirk et al. 2013, N = ln(1-D) / ln(1-P) (N = sample size, ln = natural logarithm, D = probability of detection, P = proportion of infected bees) predicts that with a sample size of five bees, pathogenic infections affecting 45% or more of the individuals within a colony would be detected with 95% probability [51]; this sample size has proven sufficient for the pathogen-specific PCR detection of highly prevalent pathogens [21–23].
Varroa destructor mite infestation level
Varroa destructor mite infestation levels for each colony were estimated using the alcohol wash technique [52]. At each sampling event, 150 adult bees were obtained from a brood-containing frame and rinsed in ethanol to dislodge attached mites into a collection jar. Mites infestation percentage was reported as the number of mites per 100 bees [52]. In total 92, samples were included in the analysis (Fig 1 and S3 Table).
RNA isolation
Five female bees per sample were homogenized together in water using beads (3 mm) and a TissueLyzer (Qiagen) at 30 Hz for 2 minutes. Samples were centrifuged for 12 minutes at 12,000 x g at 4°C to pellet debris, and RNA from supernatants was extracted using Trizol reagent (Life Technologies) according to the manufacturer’s instructions [21–23].
Reverse transcription/cDNA synthesis
cDNA synthesis reactions were performed by incubating 2 μg of total RNA extracted from honey bee samples, Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega), and 500 ng random hexamer primers (Integrated DNA Technologies) for 1 hour at 37°C, according to the manufacturer’s instructions [21,22].
Polymerase Chain Reaction (PCR)
Pathogen specific PCR was performed according to standard methods using the primers listed in S1 Table. Specifically, PCR was used to determine the prevalence of DWV, BQCV, IAPV, KBV, LSV1, LSV2, N. ceranae, and L. passim (S1 Table). In brief, each PCR included 1 μl cDNA template, combined with 10 pmol each of forward and reverse pathogen specific primers, and amplified with ChoiceTaq polymerase (Denville) according to the manufacturer’s instructions using the following cycling conditions: 95°C for 5 minutes; 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by final elongation at 72°C for 4 minutes. The PCR products were visualized using 2% agarose gel electrophoresis followed by fluorescence imaging. In addition, the PCR assays utilized in this study were verified by sequencing [22]. Positive and negative control reactions were included for all pathogen-specific PCR analyses and exhibited the expected results. IAPV was detected by PCR in only one of the samples, thus IAPV abundance was not examined by quantitative PCR.
Quantitative Polymerase Chain Reaction (qPCR)
Quantitative PCR was used to determine the relative pathogen abundance in each sample. Specifically, qPCR was used to determine the abundance of DWV, BQCV, IAPV, KBV, LSV1, LSV2, N. ceranae, and L. passim (S1 Table). All qPCR reactions were performed in triplicate with a CFX Connect Real Time instrument (BioRad) and the following reaction conditions: 2 μL of cDNA template in 20 μL reactions containing 1X ChoiceTaq Mastermix (Denville), 0.4 μM each forward and reverse primer, 1X SYBR Green (Life Technologies), and 3 mM MgCl2. The qPCR thermo-profile consisted of a single pre-incubation 95°C (1 minute), 40 cycles of 95°C (10 seconds), 58°C (20 seconds), and 72°C (15 seconds). Plasmid standards, containing from 109 to 103 copies per reaction, were used as qPCR templates to assess primer efficiency and quantify the relative abundance of each pathogen. The linear standard equation for primer efficiency of each primer set, which was generated by plotting the crossing point (Cp) versus the log10 of the initial plasmid copy number, is as follows: BQCV: y = -3.7336x+ 42.849, R2 = 0.996; DWV: y = -3.443x + 41.277, R2 = 0.99958; LSV1: y = -3.1994x + 38.71, R2 = 0.982, LSV2: y = -3.8147x + 44.805, R2 = 0.980; KBV: y = -3.4505x + 40.099, R^2 = 0.99927, L. passim: y = -3.4825x + 41.025, R2 = 0.98588 and N. ceranae: y = -3.9656 + 42.124, R2 = 0.9961; the minimum qPCR detection levels were ≤ 1,000 copies per sample. In addition, qPCR of a host encoded gene, Apis m. Rpl8, was performed using 2 μL cDNA template on each qPCR plate to ensure consistency and cDNA quality. qPCR products were analyzed by melting point analysis and 2% agarose gel electrophoresis. In addition, the qPCR assays utilized in this study were verified by sequencing [22]. An estimate of the number of viral RNA copies per bee can be obtained by multiplying the reported qPCR copy number values by 25. This estimate is based on the following: typical RNA yield was approximately 50 μg per bee, each qPCR reaction was performed on cDNA generated from 2 μg RNA, therefore each well represents 1/25th of an individual bee [21].
Statistical analysis of PCR data
For this study, we use “pathogen prevalence” to refer to the sum of the number of pathogens detected by PCR out of seven total; an additional pathogen was counted (+1) when Varroa mite infestation surpassed 3%. A one-way ANOVA was used to compare pathogen prevalence between colonies with differential colony health ratings as they were graded at the time of sampling (i.e., dead, strong, average, and weak), and to compare pathogen prevalence between sampling events (i.e., before, during, after 1, after 2, and after 3 almond pollination).
Statistical analysis of qPCR data with multiple linear regression
The associations between pathogen abundance and other monitored factors, including colony strength, mite count, and beekeeping operation were evaluated for each pathogen using multiple linear regression [22]. The natural logarithm (ln) was used to transform pathogen abundance (as determined by qPCR) to normalize the response variable; 1 was added to each observation since some observations had 0 total abundance. The confounding effects of seasonality on pathogen abundance was removed by using the residuals of a linear regression evaluating the relationship between pathogen abundance and the “day of year” of sampling as the response variable in the multiple linear regression models. As a result, models reported associations between the residuals of pathogen abundance after accounting for the effects of pathogen seasonality (“residual abundance”), and colony strength rating, beekeeping operation, and mite count. Most colonies were measured multiple times in the 92 samples; therefore, a random effect for individual colony was included in the models to account for repeated measures. Estimated models for each pathogen are described as:
Where ln(yi) represents the residuals of pathogen abundance in response to day of year of sampling for each pathogen, β0 is the estimate of the model intercept, β1 is the estimate of the change in pathogen abundance of Operation 2 compared to Operation 1, β2 is the estimate of the change in pathogen abundance for each unit increase in frame count, and β3 is the estimate of the change in pathogen abundance for each unit increase in mite count. The variable γi(j) represents a random effect for colony i sampled j times, and εi represents the residual error from the model. Coefficients (est) of the estimated linear model describe the magnitude and direction of associations between the residual abundance of individual pathogens (i.e., DWV, BQCV, KBV, LSV1, LSV2, L. passim, and N. ceranae) and other monitored factors throughout 2014.
Statistical analysis of Varroa destructor mite infestation with binomial regression
A generalized linear mixed effects model with a binomial family distribution and random effect for individual colony was used to estimate the odds that a colony will surpass the threshold of 3% mite infestation, after which treatment is recommended, in response to the date of sampling. Colonies were assigned a value of “1” when mite infestation surpassed 3%, and were assigned a value of “0” when Varroa mite infestation was less than 3%. The odds that a colony would surpass the 3% mite infestation threshold were plotted in response to the day of year and a best-fit line was fit to the data.
Multidimensional statistical analysis of pathogen compositions
We defined the pathogen composition of a honey bee colony as the abundance of seven pathogens (DWV, BQCV, LSV1, LSV2, KBV, L. passim, and N. ceranae) relative to all other samples. We used the Bray-Curtis index to create a dissimilarity matrix using the natural logarithm (ln) of pathogen abundance of seven pathogens. The relative composition of pathogens was visualized using nonmetric Multidimensional Scaling (NMDS). Relative differences in colony pathogen compositions were visualized using the five discrete categories of sampling events; January (California, before), March (California, during), June (Minnesota, after 1), August (Minnesota, after 2), and September (Minnesota, after 3).
We used a permutational analysis of variance (PERMANOVA) to test the ability of sampling event, mite count, and colony health to explain variation within the NMDS plot [53]. The main effects as well as an interaction between sampling event and mite count were included in the reported PERMANOVA model. Colony health rating was included as a main effect in a separate PERMANOVA model, but was not considered a significant predictor. Additionally, we described any differences in the variability of pathogen composition by sampling event using an analysis of multivariate homogeneity of group dispersions (betadisper) [53]. Furthermore, we used a similarity percentage (SIMPER) analysis to describe the percent dissimilarity explained by each pathogen in comparisons between consecutive sampling events [53].
Statistical software
All statistical analysis was performed using the software “R” (R version 3.1.3, “Smooth Sidewalk”) [54]. Generalized linear models were conducted using the nlme package [55]. NMDS plots were generated using the labdsv package while all other multivariate analyses were performed using the vegan and labdsv packages [53,56].
Results and discussion
Honey bee colony monitoring and pathogen diagnostics
Commercially managed honey bee colonies that were transported from Minnesota to California for almond pollination were monitored from January 2014 to January 2015 (Fig 1) [50]. At each sampling event, colony health, using colony population size as a proxy for health, was monitored and samples of live honey bees were obtained for pathogen diagnostics, including Varroa destructor mite infestation levels and the prevalence and abundance of five commonly occurring viruses (i.e., BQCV, DWV, LSV1, LSV2, and KBV), and two eukaryotic pathogens (the trypanosomatid Lotmaria passim and the microsporidial pathogen Nosema ceranae) using pathogen specific PCR and qPCR, respectively [22,23,26,50]. Israeli acute paralysis virus (IAPV) was only detected in one sample using PCR, thus it was not included in further analysis. In this work, we use “pathogen prevalence” to indicate the number of different pathogens detected in a sample and “pathogen abundance” as the estimated number of relative RNA transcripts for each pathogen using RT-qPCR (S3 Table). Furthermore, we use “pathogen composition” to refer to the abundance of all co-infecting pathogens in a sample relative to all other samples. Therefore, both pathogen prevalence and abundance affect the pathogen composition of each colony throughout the course of the study.
Pathogen prevalence and colony health
To examine the relationship between pathogen prevalence and colony health, the total number of pathogens detected in each sample, using PCR results for seven pathogens (i.e. DWV, BQCV, LSV1, LSV2, KBV, L. passim, and N. ceranae), and V. destructor mite infestation when levels were above the recommended treatment threshold of 3% (i.e., 3 mites per 100 bees) was averaged by colony health rating (i.e., weak, average, strong, or dead) (S3 Table) [7,52,57,58]. Mean pathogen prevalence varied by colony health rating (ANOVA, F3,88 = 3.05, p-value = 0.03). Specifically, samples from dead colonies averaged 6.50 pathogens (n = 2, SE +/- 1.50), weak colonies 5.25 pathogens (n = 8, SE +/- 0.38), average colonies 5.38 pathogens (n = 34 SE +/- 0.18), and strong colonies 5.95 pathogens (n = 48, SE +/- 0.13) (S2 Table and S1 Fig). Strong rated colonies had higher pathogen prevalence than average rated colonies (S1 Fig). No other pairwise comparisons of mean pathogen prevalence between colony health ratings were statistically different.
The mean pathogen prevalence associated with each colony varied with the sampling date (ANOVA, F4,87 = 21.31, p-value < 0.001) (S3 and S4 Tables and S2 Fig). Specifically, colonies sampled early in the year before almond pollination averaged 5.00 pathogens (n = 28, SE +/- 0.17), and colonies sampled in March, immediately after almond pollination and thus representing the pathogen profile of colonies during the almond bloom, averaged 5.39 pathogens (n = 28, SE +/- 0.09). Colonies sampled in early summer when the colonies were in Minnesota (i.e., after 1, June) averaged 7.09 pathogens (n = 11, SE +/- 0.28), in late summer (i.e., after 2, Aug.) colonies averaged 6.43 pathogens (n = 14, SE +/- 0.13), and at the end of summer (i.e., after 3, Sept.) colonies averaged 6.00 pathogens (n = 11, SE +/- 0.23) (S4 Table and S2 Fig). Results from Tukey’s HSD post-hoc tests indicate that the mean pathogen prevalence was greater in sampling events later in the year, when the colonies were located in Minnesota (after 1, 2, and 3) compared to sampling events in January and March, before and during almond pollination, respectively, when the colonies were located in California (S2 Fig).
Longitudinal monitoring of pathogen composition
To examine the relationships between pathogen abundance and other monitored factors (i.e., colony health, sample date, percent mite infestation, and beekeeping operation) we performed mite counts at each sampling event and utilized qPCR to quantify the relative abundance of seven pathogens (i.e. DWV, BQCV, LSV1, LSV2, KBV, L. passim, and N. ceranae) (S3 Table). Quantitative PCR values ranged from 0 to 6.7x108 per sample (S3 Table). Overall, longitudinal assessment of the abundance of common honey bee pathogens at the colony level from January to September 2014 revealed that individual colonies, both within a single beekeeping operation and between two typically managed commercial beekeeping operations, varied from the beginning of the monitoring period in January, until the end of the monitoring period in September (Fig 2 and S3–S9 Figs).
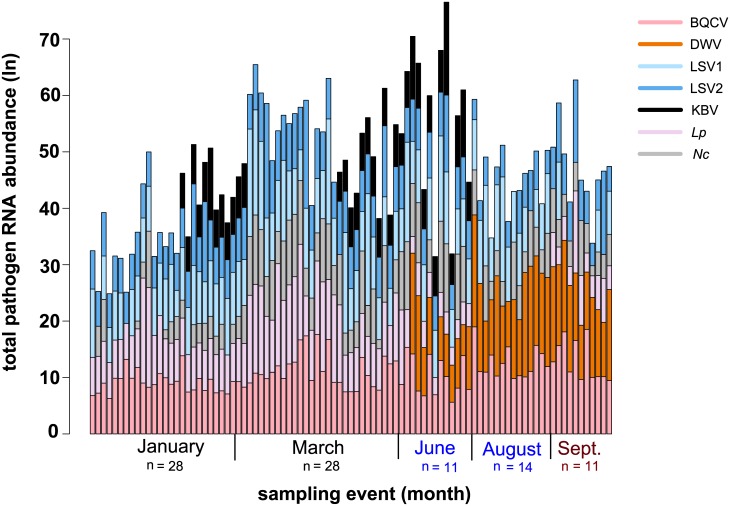
Fig 2. Longitudinal assessment of honey bee pathogen abundance at the colony level.
Spatiotemporal change of the pathogen composition of individual honey bee colonies is designated by each vertical bar. In this sample cohort, the most prominent changes are attributed to the detection of DWV and reduced abundance of L. passim (Lp) in June, August, and September. Quantitative PCR was used to determine the relative pathogen abundance (S3 Table). The natural log transformed data for each pathogen, BQCV (pink), DWV (orange), Lp (light pink), Nc (gray), LSV1 (light blue), LSV2 (dark blue), and KBV (black), is represented by the height of each segment of the vertical bar representing a sample from an individual colony (y-axis); the height of the bar represents the total pathogen abundance for each colony over the course of the study (January to September 2014, x-axis).
To investigate if weak and strong colonies had different pathogen compositions, we quantified the differences in pathogen prevalence and abundances between honey bee samples using a Bray-Curtis dissimilarity matrix. The pathogen composition of each sample relative to all other samples were visualized using a non-metric multidimensional scaling (NMDS) plot (Fig 3). Results from a PERMANOVA analysis indicated that sampling date explained the most variation in pathogen compositions (Fig 3 and Table 1, R2 = 0.61, p < 0.01), followed by levels of Varroa destructor mite infestation (Table 1, R2 = 0.01, p = 0.05), indicating pathogen composition varies seasonally and with changing levels of mite abundance [53]. Unexpectedly, pathogen composition did not differ between colony health ratings (PERMANOVA, F3,83 = 1.467, p = 0.14, R2 = 0.018). To identify the specific pathogens that contributed the most to the changes in pathogen composition between sampling events, a SIMPER analysis was used to calculate the contribution of each pathogen to the dissimilarity of pathogen compositions between consecutive sampling events (Table 2) [53]. Differences in pathogen composition of honey bee colonies between consecutive sampling events was not consistently explained by a single pathogen, but varied for each comparison, highlighting the dynamic nature of honey bee colony pathogen composition. DWV contributed the most to the differences between pathogen compositions of honey bee colonies when the mean difference in V. destructor mite infestation was greatest (Table 3), likely due to the role of mites in DWV transmission. Results from a multivariate homogeneity of group dispersions analysis (betadisper) indicated that the dispersion of pathogen composition among sampling events did not change (F4,87 = 0.27, p-value = 0.90) [53]. Together, this analysis indicated that the composition of honey bee associated pathogens is most influenced by sampling date and the level of mite infestation.
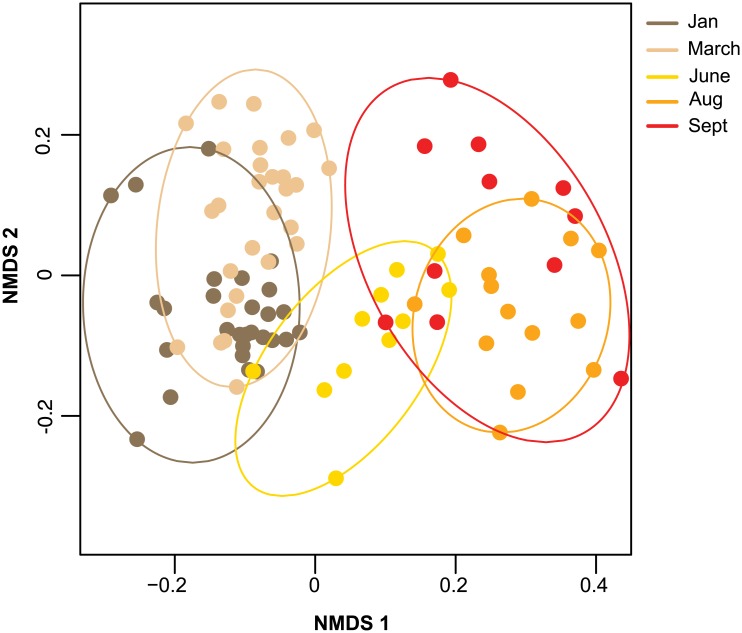
Fig 3. Relative pathogen composition of honey bee colonies visualized by sample event.
Pathogen compositions of honey bee colonies form unique and defined clusters according to the month they were sampled (i.e., January—dark brown, March—light brown, June—yellow, August—orange, or September—red). The position of each point indicates the pathogen composition of each sample relative to all other samples (i.e. samples with more similar pathogen compositions are closer), calculated using a Bray-Curtis dissimilarity and plotted on a non-metric multidimensional scaling (NMDS) plot with an associated stress value of (0.197); the results from a permutational analysis of variance (PERMANOVA) in Table 1 indicated that the sampling event explained the most amount of variance in the pathogen composition.
Table 1. Analysis of variance of pathogen composition.
The relative pathogen composition of honey bee samples, based on all pathogen abundances, is primarily explained by the date the sample was obtained (or “day of year”) and level of Varroa mite infestation. Results from a permutational analysis of variance (PERMANOVA) indicate that the day of year (R2 = 0.61), followed by the percent mite infestation (R2 = 0.01) explained the most variation in the relative pathogen composition of all honey bee samples; factors contributing significantly to the relative pathogen composition are indicated by p-values ≤0.05 in bold.
| Source | d.f. | F-Statistic | R2 | p-value |
|---|---|---|---|---|
| day of year | 4 | 34.93 | 0.61 | <0.01 |
| mite infestation (%) | 1 | 2.69 | 0.01 | 0.05 |
| sampling event x mite infestation (%) | 4 | 0.81 | 0.01 | 0.63 |
| residuals | 83 | - | 0.36 | |
| Total | 92 | - | 1.00 |
Table 2. Similarity percentage analysis of the relative pathogen composition between consecutive sampling events.
The greatest differences in honey bee colony pathogen composition was explained by changes in abundance of a different pathogen between consecutive sampling events. The percent difference attributed to each pathogen between consecutive sampling events was calculated using a similarity percentage (SIMPER) analysis of a Bray-Curtis dissimilarity matrix. The cumulative difference of the top three pathogens in comparisons between consecutive sampling events is reported as a percentage. The difference in percent mite infestation was calculated in comparisons between consecutive sampling events. When the change in mean mite infestation was greatest, DWV accounted for the most difference in honey bee colony pathogen composition.
| Sampling Event | Pathogen1 | Pathogen 2 | Pathogen 3 | Cumulative Dissimilarity (%) | Δ mean mite infestation (%) |
|---|---|---|---|---|---|
| Jan-Mar | Nc | Lp | LSV1 | 60.0 | 0.02 |
| Mar-Jun | DWV | Lp | KBV | 58.0 | 6.19 |
| Jun-Aug | KBV | DWV | Nc | 57.0 | 3.01 |
| Aug-Sept | LSV1 | Lp | LSV2 | 63.0 | -5.09 |
Table 3. Pathogen abundance model summaries.
Multiple linear regression models were utilized to identify associations between colony-level residual pathogen abundance and the factors monitored in this study (i.e., colony health, percent mite infestation, and beekeeping operation) throughout the monitoring period from January to September 2014. Residual pathogen abundance was used as the response variable in the models to remove the confounding effect of seasonality on pathogen abundance in order to investigate associations between residual pathogen abundance and other covariates. Associations between the residual abundance of each pathogen assayed in this sample cohort and other covariates are denoted by the magnitude and direction of the estimate listed in the table. Operation 2 estimates the residual pathogen abundance in comparison to Operation 1, frame count was used as a continuous variable as an estimate of colony health, and mite infestation (%) represents the number of mites counted per 100 bees. The standard error, t-value, and p-value for each estimate of coefficients (est.) within the final model are provided; p-values <0.05 are indicative of significant associations (bold).
| BQCV | estimate | SE (+/-) | t-value | p-value |
| intercept | 0.47 | 0.93 | 0.50 | 0.612 |
| operation 2 | -1.51 | 0.68 | -2.23 | 0.03 |
| frame count | 0.01 | 0.07 | 0.16 | 0.87 |
| mite infestation (%) | 0.06 | 0.06 | 1.06 | 0.29 |
| DWV | estimate | SE (+/-) | t-value | p-value |
| intercept | 0.42 | 0.77 | 0.55 | 0.59 |
| operation 2 | -0.37 | 0.52 | -0.71 | 0.48 |
| frame count | -0.09 | 0.06 | -1.49 | 0.15 |
| mite infestation (%) | 0.23 | 0.05 | 4.84 | <0.001 |
| LSV1 | estimate | SE (+/-) | t-value | p-value |
| intercept | -1.52 | 1.38 | -1.10 | 0.27 |
| operation 2 | 2.27 | 0.95 | 2.38 | 0.02 |
| frame count | 0.03 | 0.10 | -0.33 | 0.74 |
| mite infestation (%) | -0.02 | 0.08 | -0.23 | 0.82 |
| LSV2 | estimate | SE (+/-) | t-value | p-value |
| intercept | -0.54 | 0.98 | -0.54 | 0.59 |
| operation 2 | -0.41 | 0.68 | -0.61 | 0.55 |
| frame count | 0.09 | 0.08 | 1.19 | 0.24 |
| mite infestation (%) | -0.07 | 0.06 | -1.14 | 0.26 |
| KBV | estimate | SE (+/-) | t-value | p-value |
| intercept | -1.59 | 0.82 | 1.94 | 0.06 |
| operation 2 | 5.73 | 0.56 | -1.95 | <0.001 |
| frame count | -0.11 | 0.06 | -1.74 | 0.09 |
| mite infestation (%) | -0.09 | 0.05 | -1.91 | 0.06 |
| L. passim | estimate | SE (+/-) | t-value | p-value |
| intercept | -0.53 | 1.02 | -0.52 | 0.61 |
| operation 2 | -3.54 | 0.70 | -5.07 | <0.001 |
| frame count | 0.23 | 0.08 | 2.94 | 0.005 |
| mite infestation (%) | -0.01 | 0.06 | -0.19 | 0.84 |
| N. ceranae | estimate | SE (+/-) | t-value | p-value |
| intercept | -1.92 | 1.22 | -1.57 | 0.12 |
| operation 2 | 0.20 | 0.83 | 0.24 | 0.81 |
| frame count | 0.18 | 0.09 | 1.99 | 0.05 |
| mite infestation (%) | -0.06 | 0.07 | -0.75 | 0.46 |
| total abundance | estimate | SE (+/-) | t-value | p-value |
| intercept | -0.33 | 1.02 | -0.33 | 0.75 |
| operation 2 | -1.49 | 0.73 | -2.05 | 0.05 |
| frame count | 0.08 | 0.08 | 1.00 | 0.32 |
| mite infestation (%) | 0.10 | 0.06 | 1.64 | 0.11 |
| odds surpassing Varroa threshold | estimate | SE (+/-) | t-value | p-value |
| Intercept | -51.02 | 14.35 | -3.56 | <0.001 |
| day of year | 0.26 | 0.07 | 3.66 | <0.001 |
Varroa destructor and colony health
V. destructor mites are a major contributor to honey bee colony losses [10,25,44,57,59].
V. destructor mite infestation increased over the course of this study. Specifically, honey bee samples from colonies early in the year, when the colonies were located in California, harbored a mean mite infestation of 0.45% (mean = 0.45, SE +/- 0.09, n = 56) (S3 Table). Whereas, honey bee samples obtained from colonies later in the year, when the colonies were located in Minnesota after almond pollination (i.e. after1, after2, and after3), harbored a 7.17% mean mite infestation (mean = 7.17, SE+/- 1.21, n = 36) (S3 Table). To evaluate the risk Varroa mites pose to colonies throughout the sampling period, results from a generalized linear mixed effects model with a binomial family distribution demonstrate that the odds of a colony exceeding the recommended treatment threshold level of 3% Varroa mite infestation is e0.26 (1.3) times higher with each increase in the day of the year (log-odds of surpassing threshold, est = 0.23) (Fig 4 and Table 3). These results indicate that honey bee colonies within this sample cohort frequently accumulated higher levels of mites toward the end of the pathogen monitoring period (Fig 4), consistent with the typical mite population growth pattern that tracks the seasonal colony buildup and decline over the course of a year.
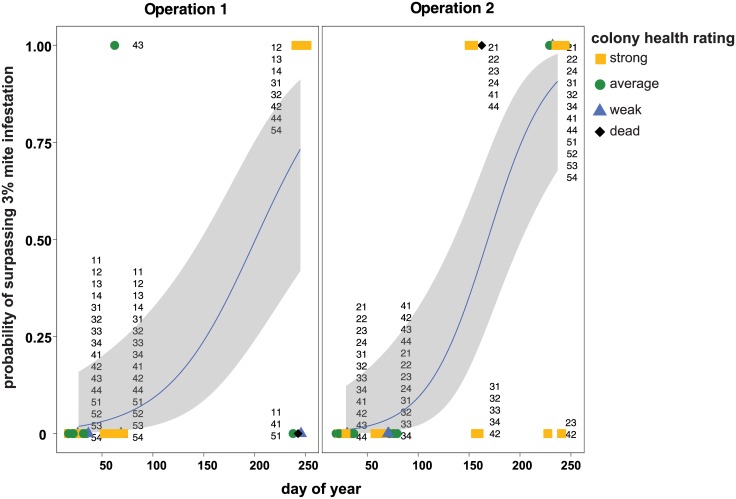
Fig 4. The probability of honey bee colonies accumulating Varroa destructor mite densities above the threshold recommended for treatment is greater later in the year.
Honey bee colonies have a high probability of accumulating levels of Varroa destructor mites that surpass the threshold for recommended treatment (i.e., 3%) by the end of the pathogen monitoring period in both beekeeping operations. Results from a generalized linear mixed effects model with a binomial family distribution indicate that by the end of the sampling period, colonies had a 99.0% chance of crossing the recommended treatment threshold. Since mite count data differed between beekeeping operations, binomial regression results were plotted independently. Mite count data obtained from honey bee samples collected from colonies rated dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square) are shown with their unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony. A best-fit line (blue) with odds-estimates surrounded by upper and lower standard error estimates (gray) depicts the odds of a colony surpassing the recommended treatment threshold (y-axis), which increases with the day of the year (x-axis).
In total, 21 colonies in this sample cohort died by the end of the sampling period in January 2015. From January 2014 to January 2015, 22 of the monitored colonies surpassed the 3% mite infestation threshold, and of those colonies that died, 40.9% (9/22) of colonies surpassed the 3% mite infestation threshold by the end of the sampling period. However, the percentage of colonies that had at some point in the study surpassed the 3% mite infestation threshold and died differed by beekeeping operation; Beekeeping Operation 1 experienced 22.2% loss (i.e., 2 of 9 colonies with > 3% mite infestation) and Beekeeping Operation 2 experienced 53.8% loss (i.e., 7 of 13 colonies with > 3% mite infestation) (S3 Table). It is notable that both beekeeping operations had numerous colonies with high levels of mite infestation, despite application of several anti-mite treatments including amitraz, oxalic acid, and formic acid over the course of the sampling period (see Methods).
Association of pathogen abundance and monitored factors, after accounting for sample date
Individual pathogen abundance was consistently affected by the sample date (“day of year”) (S3–S9 Figs), therefore we further examined the relationship between pathogen abundance and other factors (e.g., beekeeping operation, colony health, and mite infestation) after removing the confounding effects of sample date. In order to remove the confounding effects of sample date on pathogen abundance, we examined the relationship between the residuals of a linear regression of pathogen abundance in response to “day of year” (i.e., residual abundance), and used residual abundance as the response variable in multiple linear regression to evaluate associations with residual abundance and beekeeping operation, mite infestation percentage, and colony health, which was represented in the models as frame count (Table 3).
Concurrent with increased V. destructor mite levels were increased DWV prevalence and abundance (Figs 2 and 5). All the honey bee samples in this sample cohort (n = 92) were tested for the presence of DWV using PCR (S3 Table). Interestingly, none of the samples collected in January and March when the colonies were located in California (n = 56) were DWV positive, whereas 96.4% of samples obtained from colonies located in Minnesota (n = 36) were positive for DWV. Quantitative PCR was utilized to determine the relative DWV abundance in each sample (n = 92), which in general increased over the course of the monitoring period (Fig 5 and S3 Table). Multiple linear regression was used to investigate associations between residual DWV abundance with the predictor of interest, frame count (i.e., colony strength), while also accounting for beekeeping operation and level of mite infestation, indicating that median residual DWV abundance increased by e0.23 (126%) for each unit increase in mite infestation after accounting for beekeeping operation and frame count (Table 3). These data support previous results describing the relationship between mite infestation and DWV abundance [7,14,45].
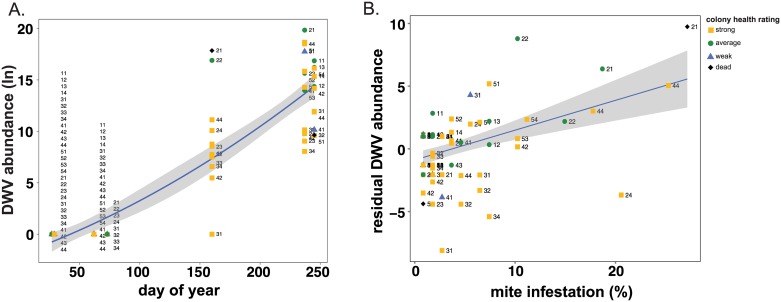
Fig 5. DWV abundance increased from January to September 2014.
DWV abundance was greatest in honey bee samples with higher mite infestation levels, obtained later in the year (June through September). A. The natural log transformed values of the relative DWV RNA abundance as determined by qPCR (y-axis) analysis of honey bee samples obtained from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)); the date of sample collection (i.e. day of year, x-axis). The best-fit line (blue), surrounded by upper and lower standard error estimates (gray), indicates that DWV abundance increased at an exponential rate across the longitudinal monitoring period. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period. B. The residual abundance of the natural log transformed values of DWV RNA abundance (y-axis) increases as the level of mite infestation (x-axis) increases.
KBV was only detected in Operation 2, and as a result residual abundance was estimated in the model to be greater within Operation 2 when compared to Operation 1, while also accounting for frame count and mite infestation percentage (Table 3). While not significant, a negative association was observed between residual KBV abundance and levels of mite infestation (mites, est = -0.09, SE = 0.06, p-value = 0.06) (Table 3). Mites transmit both DWV and KBV, therefore the positive correlation between DWV and mite infestation was expected, whereas further investigation is required to determine the relationship between KBV and mite infestation [44,46,47].
The residual abundance patterns of other viruses did not consistently associate with the levels of Varroa destructor mite infestation. LSV2 is a member of the recently discovered and described Lake Sinai virus group, which is prevalent, abundant, and globally distributed [21–23,31,60,61]. Consistent with other studies, LSV2 abundance was higher in weak colonies, as compared to healthy colonies with greater bee populations in this sample cohort [23,28], but this observation was only statistically supported when colony health was categorized into health ratings based on frame count. The median residual LSV2 abundance was e7.39 (161970.60%) greater in “dead” colonies compared to “weak” rated colonies (t-test, t-value56 = 3.02, p = 0.004), when categorical colony health data, as opposed continuous frame count data, was used in the models, while also accounting for the effect of beekeeping operation and mite infestation. However, samples were only collected from 2 of the 22 colonies that died. Despite the close phylogenetic relationship of LSV1 and LSV2 [23], changes in LSV1 abundance were associated with monitored factors that were different from those that correlated with LSV2 abundance. Although LSV1 was similarly widespread, testing positive in 86.9% of samples (80/92) (S3 Table), median residual LSV1 abundance was e2.27 (967.94%) times greater in Operation 2 compared to Operation 1 when also accounting for frame count and mite infestation (Operation 2, est = 2.27, SE ± 0.95, p = 0.02) (Table 3) (S6 Fig).
Counterintuitively, the residual abundance of L. passim and N. ceranae increased with colony population size estimated by frame count (Table 3). The median residual abundance of L. passim increased by e0.23 (125%) with each unit increase in frames counted, and the median residual abundance of N. ceranae increased by e0.18 (119.72%) in response to each unit increase in frames counted when also accounting for the effects of beekeeping operation, and mites (Table 3). These results differ from previous studies demonstrating that N. ceranae influences co-infections of other pathogens important to reduced colony health in the US [10,28,49], or as the causative agent of colony loss in Spain [62,63].
Furthermore, we examined correlations between changes in individual colony health between time points (Δ-Frame Count) and pathogen abundance (S10 Fig). For all pathogens except KBV, there was no correlation between abundance and Δ-Frame Count independent of day of year (S10 Fig). There was a negative correlation between KBV abundance and Δ-Frame Count independent of day of year (S10 Fig).
Pathogen abundance and colony health evaluated by beekeeping operation
The abundance of each pathogen associated with samples obtained from monitored honey bee colonies managed by two commercial beekeeping operations was plotted as function of time (i.e., day of the year) (S3–S9 Figs). Together these data illustrate the similarities and differences in pathogen loads over the course of this study, and between colonies managed by different beekeeping operations, which encompasses numerous confounding variables including precise geographic location, intensities of exposures to pathogens, honey bee colony genetics, the availability and quality of bee forage, and specific management practices, all of which may impact colony health and pathogen prevalence and abundance. In this sample cohort, the abundances of BQCV, LSV1, KBV, and L. passim, and the total pathogen abundance differed between beekeeping operations (Table 3). However, there was a large gap in data between March and August for each beekeeping operation, thus this study cannot attribute differences in pathogen abundance to apiary management. In previous US based honey bee monitoring studies, the specific beekeeping operation affected pathogen abundances [22]; however, this was not observed in colonies monitored as part of the German Bee Monitoring Project [25]. Additionally, in other studies, apiaries affected by CCD had more weak colonies with higher pathogen prevalence within closer proximity to other weak colonies with high pathogen prevalence [10]. Future investigations aimed at better understanding the mechanisms and dynamics of pathogen transmission between colonies within the same apiary (e.g., via mites, foraging/pollen, robbing, beekeeper mediated) are important to developing strategies to reduce inter-colony transmission.
Pairwise pathogen interactions
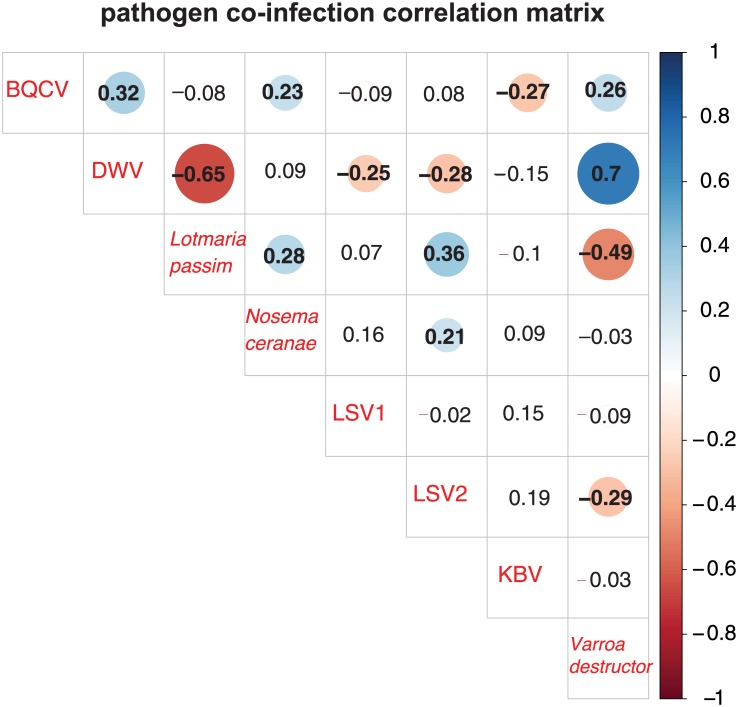
Honey bee colonies are infected by a suite of pathogens that fluctuate in prevalence and abundance over the course of a year [10,22,28,31,64]. To investigate pairwise interactions between the pathogens monitored in this study, we generated a correlation matrix illustrating the correlation coefficients (R-values) that quantify the strength and direction of the relationship between pathogen abundance data (Fig 6). The strongest positive correlation observed in this sample cohort was between V. destructor mites and DWV (R = 0.70, p-value < 0.001). DWV and L. passim (R = -0.65, p-value < 0.001), followed by V. destructor mites and L. passim (R = -0.49, p-value < 0.001), demonstrated the strongest negative correlations (Fig 6).
Fig 6. Pathogen co-infection correlation matrix.
The abundance of pathogens in co-infected colonies was analyzed by calculating the correlation coefficients for each pair-wise comparison, which are listed in each cell. Correlation coefficients (reported as r values) quantify the strength and direction of the changes in pathogen abundance between pairs of pathogens. The shaded red circles represent negative correlations and blue circles represent positive correlations, darker hues and larger circles indicate stronger correlations, and bold numbers indicate significant correlations (p-value < 0.05). DWV and Varroa destructor mites had the strongest positive correlation, while DWV and L. passim displayed the strongest negative correlation.
Conclusions
The health of commercially managed, migratory honey bee colonies in the US [7,10,21,22] and honey colonies located throughout the world [31,61,65–68] is influenced by multiple biotic and abiotic factors. In addition to pathogens, other factors including seasonal variation in colony size, foraging requirements, nutrition, and pesticide exposure all impact colony health [10,35,38,69,70]. Therefore, temporal monitoring projects that emphasize robust sampling designs (e.g., consistent monthly samples obtained on a specific date) and include large sample sizes and multiple monitored factors, are required to fully describe both general and acute threats to honey bee colonies throughout the year and in the context of pollination of specific crops. Additionally, the synergistic effects of co-infecting honey bee pathogens [35,64], which can impact the prevalence and abundance of other pathogens [7,28,31,49], must be investigated from a multidimensional approach to study the effects of pathogens on honey bee colony health. The combination of sampling individual honey bee colonies on specific days of the year and across multiple sites, and employment of statistical methods aimed at integrating and interpreting multifactorial data sets, will further our understanding of the factors with the greatest influence on colony losses and may result in the development of strategies to limit future losses.
Supporting information
(PDF)
Pathogen prevalence varied significantly between strong and average colony health ratings. Honey bee samples obtained from dead, weak, average, and strong colonies were tested for the presence of seven pathogens, including five viruses (i.e., DWV, BQCV, LSV1, LSV2, KBV), L. passim, and N. ceranae using PCR, and for Varroa destructor mites, which were counted as a pathogen when infestation levels were above the recommended treatment threshold of 3%. Total pathogen prevalence was determined by summing the number of pathogens detected in each sample. Honey bee colony population size was used as a proxy for colony health by counting the number of frames more than 2/3 covered with bees (i.e. weak < 7 frames, average = 7–12 frames, strong >12 frames covered with bees). Included in the table are the mean number of pathogens per colony strength rating, the standard error estimate of the mean, and the number of colonies per colony health rating within this cohort. This sample cohort was comprised of 52.2% strong, 37% average, 8.6% weak, and 2.2% dead colonies.
(PDF)
Honey bee colonies from two Minnesota-based commercial beekeeping operations that transport honey bee colonies to California for almond pollination were monitored for two years from January 2014 to January 2015. At the onset of the study in January 2014, beekeepers identified 12–16 queen-right colonies that successfully overwintered (n = 28), of which two died throughout the course of the study and were replaced by three novel colonies bringing the total unique colonies up to 31 throughout the course of the study (n = 31); repeated sampling of colonies resulted in 93 total samples. Honey bee colony population size was used as a proxy for colony health by counting the number of frames more than 2/3 covered with bees (i.e. weak < 7 frames, average = 7–12 frames, strong >12 frames covered with bees). Pathogen diagnostics was performed by PCR (1 = positive detection, 0 = not detected, NA—not assessed or no sample) and quantitative PCR (qPCR). Relative RNA equivalents were determined by qPCR and were natural log transformed for statistical analyses.
(XLS)
Pathogen prevalence varied significantly by sampling event. Honey bee samples were obtained from two Minnesota based, commercially managed honey bee colonies, twice when colonies were located in California before and immediately after almond pollination and thus reflect the pathogen prevalence during the almond bloom, and three times after the almond pollination event when colonies were located in Minnesota. Pathogen prevalence was measured by totaling the number of pathogens detected via PCR in each sample. Included are the average number of pathogens per sampling event, the standard error estimate of the mean, and the number of colonies per colony health rating within this cohort.
(PDF)
Mean pathogen prevalence is higher in samples from strong rated colonies, than in samples from average rated colonies. The number of pathogens detected by PCR, and mites (when infestation percentage surpassed 3 mites per 100 bees) were summed for each sample and averaged (y-axis) for each colony health rating (x-axis). Colony health was rated using colony size as a proxy, where colonies with <7 frames covered with bees were rated weak (n = 8, blue), 7–12 frames were rated average (n = 34), >12 frames were rated strong (n = 48, gold), or 0 frames were rated dead (n = 2, black). Points represent raw data, and center bars represent the mean colony pathogen prevalence, which are surrounded by the upper and lower standard error estimates. Significant differences between mean pathogen prevalence of colony health ratings was calculated using Tukey’s HSD post-hoc test (p-value < 0.05).
(EPS)
Colonies acquired the highest mean number of pathogens by the first sampling event after almond pollination. The number of pathogens detected by PCR, and mites (when infestation percentage surpassed 3 mites per 100 bees) were summed in each sample and averaged (y-axis) for each sampling event (x-axis). Sampling events were conducted once before (n = 28, dark brown), once during (n = 28, light brown), and three times after California almond pollination (n = 11 yellow, n = 14 orange, and n = 11 red). Points represent raw data, and center bars represent the mean pathogen prevalence, which are surrounded by the upper and lower standard error estimates. Significant differences between the mean colony-level pathogen prevalence between sampling events were calculated using Tukey’s HSD post-hoc test (p-value < 0.05). The mean pathogen prevalence was greater in sampling events carried out after almond pollination when the colonies were located in Minnesota (after 1, 2, and 3) compared to sampling events before and during almond pollination when the colonies were located in California (with the exception of the pairwise comparison between during and after 3).
(EPS)
Total pathogen abundance changed throughout the monitoring period, and was different between beekeepers. The natural log transformed values of individual pathogens measured by qPCR were summed (y-axis) and plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies rated dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
KBV abundance varied across the monitoring period in beekeeping operation 2, but was never detected in colonies from beekeeping operation 1. The natural log transformed values of the relative KBV abundance as determined by qPCR (y-axis) in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). KBV abundance in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on KBV abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
LSV2 abundance was weakly correlated with date of sampling, but was greater in dead colonies. The natural log transformed values of the relative LSV2 RNA abundance as determined by qPCR (y-axis in honey bee samples were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
LSV1 abundance changed throughout the monitoring period, and was different between beekeeping operations. The natural log transformed values of the relative LSV1 RNA abundance as determined by qPCR (y-axis in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
N. ceranae abundance changed throughout the monitoring period and was predicted to be greater in strong rated colonies, compared to weak rated colonies. The natural log transformed values of the relative N. ceranae abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
L. passim abundance was predicted to change throughout the monitoring period, but the abundance was greater in beekeeping Operation 1, than beekeeping Operation 2. The natural log transformed values of the relative L. passim abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). L. passim abundance in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on L. passim abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
BQCV abundance changed throughout the monitoring period, and levels of abundance were higher in beekeeping Operation 2, than in beekeeping Operation 1. The natural log transformed values of the relative BQCV abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). BQCV abundances in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on BQCV abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
The residuals of the change in frame count (used as a proxy for changes in colony health rating), after accounting for the sampling date (i.e., “day of year”) (y-axis) were plotted relative to the natural log transformed values of the relative pathogen RNA abundance (x-axis) for (A.) DWV (B.) LSV1, (C.) LSV2, (D.) BQCV, (E.) KBV, (F.) N. ceranae, and (G.) L. passim. Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). The best-fit line (blue) surrounded by upper and lower standard error estimates (gray) indicates trends in the changes in frame count relative to pathogen abundance. After accounting for changes in frame counts over time, the only significant result was a negative relationship between KBV abundance and changes in frame count. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph.
(EPS)
Acknowledgments
This research was supported by the National Honey Board and in addition the Flenniken Laboratory is supported by the National Sciences Foundation CAREER Program, the United States Department of Agriculture National Institute of Food and Agriculture, Agriculture and Food Research Initiative (USDA-NIFA-AFRI) Program, Montana Department of Agriculture Specialty Crop Block Grant Program, the National Institutes of Health IDeA Program COBRE grant GM110732, National Science Foundation EPSCoR NSF-IIA-1443108, Hatch Multistate Funding (NC-1173), Project Apis m., the Montana State Beekeepers Association, Montana State University, and the Montana State University Agricultural Experiment Station. We would like to thank members of the Flenniken laboratory at Montana State University (Dr. Laura Brutscher, Dr. Katie Daughenbaugh, and Alex McMenamin), Geoffrey W. Marcy (Pesticide Research Institute), and Dr. Marie Pizzorno (Bucknell University) for reviewing this manuscript prior to publication and Dr. Kezia Manlove (USGS Washington State University) for consultation on statistical analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Honey Board. In addition, the Flenniken Laboratory is supported by the National Sciences Foundation CAREER Program, the United States Department of Agriculture National Institute of Food and Agriculture, Agriculture and Food Research Initiative (USDA-NIFA-AFRI) Program, Montana Department of Agriculture Specialty Crop Block Grant Program, the National Institutes of Health IDeA Program COBRE grant GM110732, National Science Foundation EPSCoR NSF-IIA-1443108, Hatch Multistate Funding (NC-1173), Project Apis m., the Montana State Beekeepers Association, Montana State University, and the Montana State University Agricultural Experiment Station. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Calderone NW (2012) Insect Pollinated Crops, Insect Pollinators and US Agriculture: Trend Analysis of Aggregate Data for the Period 1992–2009. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gallai N, Salles JM, Settele J, Vaissiere BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics 68: 810–821. [Google Scholar]
- 3.(2014) California ABO Almond Almanac.
- 4.Garibaldi LA, Steffan-Dewenter I, Winfree R, Aizen MA, Bommarco R, Cunningham SA, et al. (2013) Wild Pollinators Enhance Fruit Set of Crops Regardless of Honey Bee Abundance. Science 339: 1608–1611. doi: 10.1126/science.1230200 [DOI] [PubMed] [Google Scholar]
- 5.Ollerton J, Winfree R, Tarrant S (2011) How many flowering plants are pollinated by animals? Oikos 120: 321–326. [Google Scholar]
- 6.Lee KV, Steinhauer N, Rennich K, Wilson ME, Tarpy DR, Caron DM, et al. (2015) A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie 46: 292–305. [Google Scholar]
- 7.Traynor KS, Rennich K, Forsgren E, Rose R, Pettis J, Kunkel G, et al. (2016) Multiyear survey targeting disease incidence in US honey bees. Apidologie 47: 325–347. [Google Scholar]
- 8.NASS U (2016) Honey Bee Colonies.
- 9.vanEngelsdorp D, Hayes J, Underwood RM, Pettis J (2008) A Survey of Honey Bee Colony Losses in the US, Fall 2007 to Spring 2008. Plos One 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.vanEngelsdorp D, Evans JD, Saegerman C, Mullin C, Haubruge E, Nguyen BK, et al. (2009) Colony Collapse Disorder: A Descriptive Study. Plos One 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.vanEngelsdorp D, Caron D, Hayes J, Underwood R, Henson M, Rennich K, et al. (2012) A national survey of managed honey bee 2010–11 winter colony losses in the USA: results from the Bee Informed Partnership. Journal of Apicultural Research 51: 115–124. [Google Scholar]
- 12.Robinson GE, Evans JD, Maleszka R, Robertson HM, Weaver DB, Worley K, et al. (2006) Sweetness and light: illuminating the honey bee genome. Insect Mol Biol 15: 535–539. doi: 10.1111/j.1365-2583.2006.00698.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryabov EV, Fannon JM, Moore JD, Wood GR, Evans DJ (2016) The Iflaviruses Sacbrood virus and Deformed wing virus evoke different transcriptional responses in the honeybee which may facilitate their horizontal or vertical transmission. PeerJ 4: e1591 doi: 10.7717/peerj.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mordecai GJ, Wilfert L, Martin SJ, Jones IM, Schroeder DC (2016) Diversity in a honey bee pathogen: first report of a third master variant of the Deformed Wing Virus quasispecies. Isme Journal 10: 1264–1273. doi: 10.1038/ismej.2015.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smart M, Pettis J, Rice N, Browning Z, Spivak M (2016) Linking Measures of Colony and Individual Honey Bee Health to Survival among Apiaries Exposed to Varying Agricultural Land Use. Plos One 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danka RG, De Guzman LI, Rinderer TE, Sylvester HA, Wagener CM, Bourgeois Al, et al. (2012) Functionality of Varroa-Resistant Honey Bees (Hymenoptera: Apidae) When Used in Migratory Beekeeping for Crop Pollination. Journal of Economic Entomology 105: 313–321. [DOI] [PubMed] [Google Scholar]
- 17.Simone-Finstrom M, Li-Byarlay H, Huang MH, Strand MK, Rueppell O, Tarpy D. (2016) Migratory management and environmental conditions affect lifespan and oxidative stress in honey bees. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsvetkov N, Samson-Robert O, Sood K, Patel H, Malena D, Gajiwala PH, et al. (2017) Chronic exposure to neonicotinoids reduces honey bee health near corn crops. Science 356: 1395–1397. doi: 10.1126/science.aam7470 [DOI] [PubMed] [Google Scholar]
- 19.COLOSS (2013) COLOSS BEEBOOK: Standard Methods in Apicultural Research. [Google Scholar]
- 20.Sagili R, Burgett D (2011) Evaluating honey bee colonies for pollination. Pacific Northwest Extension Publications. [Google Scholar]
- 21.Runckel C, Flenniken ML, Engel JC, Ruby JG, Ganem D, Andino R, et al. (2011) Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One 6: e20656 doi: 10.1371/journal.pone.0020656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavigli I, Daughenbaugh KF, Martin M, Lerch M, Banner K, Garcia E, et al. (2016) Pathogen prevalence and abundance in honey bee colonies involved in almond pollination. Apidologie 47: 251–266. doi: 10.1007/s13592-015-0395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daughenbaugh KF, Martin M, Brutscher LM, Cavigli I, Garcia E, Lavin M, et al. (2015) Honey Bee Infecting Lake Sinai Viruses. Viruses-Basel 7: 3285–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langstroth L (2016) Langstroth on the Hive and the Honey-Bee: Xist Publishing. [Google Scholar]
- 25.Genersch E, von der Ohe W, Kaatz H, Schroeder A, Otten C, Büchler R, et al. (2010) The German bee monitoring project: a long term study to understand periodically high winter losses of honey bee colonies. Apidologie 41: 332–352. [Google Scholar]
- 26.Tentcheva D, Gauthier L, Zappulla N, Dainat B, Cousserans F, et al. (2004) Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Applied and Environmental Microbiology 70: 7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YP, Pettis JS, Corona M, Chen WP, Li CJ, Spivak M, et al. (2014) Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. Plos Pathogens 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornman RS, Tarpy DR, Chen YP, Jeffreys L, Lopez D, Pettis JS, et al. (2012) Pathogen Webs in Collapsing Honey Bee Colonies. Plos One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, et al. (2007) A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283–287. doi: 10.1126/science.1146498 [DOI] [PubMed] [Google Scholar]
- 30.Seitz N, Traynor KS, Steinhauer N, Rennich K, Wilson ME, Ellis JD, et al. (2016) A national survey of managed honey bee 2014–2015 annual colony losses in the USA. Journal of Apicultural Research 54: 292–304. [Google Scholar]
- 31.Ravoet J, Maharramov J, Meeus I, De Smet L, Wenseleers T, Smagghe G, et al. (2013) Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen BK, Mignon J, Laget D, de Graaf DC, Jacobs FJ, vanEngelsdorp D, et al. (2010) Honey bee colony losses in Belgium during the 2008–9 winter. Journal of Apicultural Research 49: 337–339. [Google Scholar]
- 33.Evans JD, Schwarz RS (2011) Bees brought to their knees: microbes affecting honey bee health. Trends in Microbiology 19: 614–620. doi: 10.1016/j.tim.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 34.Brutscher LM, McMenamin AJ, Flenniken ML (2016) The Buzz about Honey Bee Viruses. Plos Pathogens 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen YP, Siede R (2007) Honey bee viruses. Advances in Virus Research, Vol 70 70: 33–80. [DOI] [PubMed] [Google Scholar]
- 36.Genersch E, Aubert M (2010) Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Veterinary Research 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Miranda JR, Cordoni G, Budge G (2010) The Acute bee paralysis virus-Kashmir bee virus-Israeli acute paralysis virus complex. J Invertebr Pathol 103 Suppl 1: S30–47. [DOI] [PubMed] [Google Scholar]
- 38.McMenamin AJ, Brutscher LM, Glenny W, Flenniken ML (2016) Abiotic and biotic factors affecting the replication and pathogenicity of bee viruses. Current Opinion in Insect Science 16: 14–21. doi: 10.1016/j.cois.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Runckel C, DeRisi J, Flenniken ML (2014) A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae. PLoS One 9: e95057 doi: 10.1371/journal.pone.0095057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz RS, Bauchan GR, Murphy CA, Ravoet J, de Graaf DC, Evans J. (2015) Characterization of Two Species of Trypanosomatidae from the Honey Bee Apis mellifera: Crithidia mellificae Langridge and McGhee, 1967 and Lotmaria passim n. gen., n. sp. Journal of Eukaryotic Microbiology 62: 567–583. doi: 10.1111/jeu.12209 [DOI] [PubMed] [Google Scholar]
- 41.Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B,et al. (2009) Genomic Analyses of the Microsporidian Nosema ceranae, an Emergent Pathogen of Honey Bees. Plos Pathogens 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genersch E (2010) American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. Journal of Invertebrate Pathology 103: S10–S19. doi: 10.1016/j.jip.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 43.Forsgren E (2010) European foulbrood in honey bees. Journal of Invertebrate Pathology 103: S5–S9. doi: 10.1016/j.jip.2009.06.016 [DOI] [PubMed] [Google Scholar]
- 44.Nazzi F, Le Conte Y (2016) Ecology of Varroa destructor, the Major Ectoparasite of the Western Honey Bee, Apis mellifera. Annual Review of Entomology, Vol 61 61: 417–432. [DOI] [PubMed] [Google Scholar]
- 45.Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, et al. (2014) A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. Plos Pathogens 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YP, Pettis JS, Evans JD, Kramer M, Feldlaufer MF (2004) Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 35: 441–448. [Google Scholar]
- 47.Shen MQ, Yang XL, Cox-Foster D, Cui LW (2005) The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 342: 141–149. doi: 10.1016/j.virol.2005.07.012 [DOI] [PubMed] [Google Scholar]
- 48.Di Prisco G, Pennacchio F, Caprio E, Boncristiani HF Jr., Evans JD, Chen Y. (2011) Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J Gen Virol 92: 151–155. doi: 10.1099/vir.0.023853-0 [DOI] [PubMed] [Google Scholar]
- 49.Nazzi F, Brown SP, Annoscia D, Del Piccolo F, Di Prisco G, Varricchio P, et al. (2012) Synergistic Parasite-Pathogen Interactions Mediated by Host Immunity Can Drive the Collapse of Honeybee Colonies. Plos Pathogens 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Susan Kegley RR, Geoff (2017) Pesticiced residues in honey bee colonies involved in 2014 California almond pollination. PLoS One. [Google Scholar]
- 51.Pirk CW, De Miranda JR, Kramer M, Murray TE, Nazzi F, Shutler D, et al. (2013) Statistical guidelines for Apis mellifera research. Journal of Apicultural Research 52: 1–24. [Google Scholar]
- 52.Lee KV, Moon RD, Burkness EC, Hutchison WD, Spivak M (2010) Practical Sampling Plans for Varroa destructor (Acari: Varroidae) in Apis mellifera (Hymenoptera: Apidae) Colonies and Apiaries. Journal of Economic Entomology 103: 1039–1050. [DOI] [PubMed] [Google Scholar]
- 53.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. (2007) The vegan package. Community ecology package 10.
- 54.Team RC (2013) R: A language and environment for statistical computing. [Google Scholar]
- 55.Pinheiro J, Bates D, DebRoy S, Sarkar D (2014) R Core Team (2014) nlme: linear and nonlinear mixed effects models. R package version 3.1–117 http://CRAN R-project org/package = nlme. [Google Scholar]
- 56.Roberts D (2007) labdsv: Ordination and multivariate analysis for ecology. R package version 1. [Google Scholar]
- 57.Currie RW, Gatien P (2006) Timing acaricide treatments to prevent Varroa destructor (Acari: Varroidae) from causing economic damage to honey bee colonies. Canadian Entomologist 138: 238–252. [Google Scholar]
- 58.Giacobino A, Molineri A, Cagnolo NB, Merke J, Orellano E, Bertozzi E, et al. (2015) Risk factors associated with failures of Varroa treatments in honey bee colonies without broodless period. Apidologie 46: 573–582. [Google Scholar]
- 59.Genersch E (2010) Honey bee pathology: current threats to honey bees and beekeeping. Applied Microbiology and Biotechnology 87: 87–97. doi: 10.1007/s00253-010-2573-8 [DOI] [PubMed] [Google Scholar]
- 60.Granberg F, Vicente-Rubiano M, Rubio-Guerri C, Karlsson OE, Kukielka D, et al. (2013) Metagenomic Detection of Viral Pathogens in Spanish Honeybees: Co- Infection by Aphid Lethal Paralysis, Israel Acute Paralysis and Lake Sinai Viruses. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cepero A, Ravoet J, Gomez-Moracho T, Bernal JL, Del Nozal MJ, et al. (2014) Holistic screening of collapsing honey bee colonies in Spain: a case study. BMC Res Notes 7: 649 doi: 10.1186/1756-0500-7-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin-Hernandez R, Meana A, Prieto L, Salvador AM, Garrido-Bailon E, et al. (2007) Outcome of colonization of Apis mellifera by Nosema ceranae. Applied and Environmental Microbiology 73: 6331–6338. doi: 10.1128/AEM.00270-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higes M, Martin-Hernandez R, Botias C, Bailon EG, Gonzalez-Porto AV, et al. (2008) How natural infection by Nosema ceranae causes honeybee colony collapse. Environmental Microbiology 10: 2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x [DOI] [PubMed] [Google Scholar]
- 64.McMenamin AJ, Genersch E (2015) Honey bee colony losses and associated viruses. Current Opinion in Insect Science 8: 121–129. [DOI] [PubMed] [Google Scholar]
- 65.van der Zee R, Pisa L, Andonov S, Brodschneider R, Charriere JD, Chlebo R, et al. (2012) Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. Journal of Apicultural Research 51: 91–114. [Google Scholar]
- 66.Berenyi O, Bakonyi T, Derakhshifar I, Koglberger H, Nowotny N (2006) Occurrence of six honeybee viruses in diseased Austrian apiaries. Applied and Environmental Microbiology 72: 2414–2420. doi: 10.1128/AEM.72.4.2414-2420.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nielsen SL, Nicolaisen M, Kryger P (2008) Incidence of acute bee paralysis virus, black queen cell virus, chronic bee paralysis virus, deformed wing virus, Kashmir bee virus and sacbrood virus in honey bees (Apis mellifera) in Denmark. Apidologie 39: 310–314. [Google Scholar]
- 68.Clermont A, Eickermann M, Kraus F, Georges C, Hoffmann L, Beyer M. (2014) A survey on some factors potentially affecting losses of managed honey bee colonies in Luxembourg over the winters 2010/2011 and 2011/2012. Journal of Apicultural Research 53: 43–56. [Google Scholar]
- 69.Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, vanEngelsdorp D, et al. (2010) RNA Viruses in Hymenopteran Pollinators: Evidence of Inter-Taxa Virus Transmission via Pollen and Potential Impact on Non-Apis Hymenopteran Species. Plos One 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Degrandi-Hoffman G, Chen YP (2010) Why Supplemental Protein Feeding Can Help Reduce Colony Losses. American Bee Journal 150: 1157–1160. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Pathogen prevalence varied significantly between strong and average colony health ratings. Honey bee samples obtained from dead, weak, average, and strong colonies were tested for the presence of seven pathogens, including five viruses (i.e., DWV, BQCV, LSV1, LSV2, KBV), L. passim, and N. ceranae using PCR, and for Varroa destructor mites, which were counted as a pathogen when infestation levels were above the recommended treatment threshold of 3%. Total pathogen prevalence was determined by summing the number of pathogens detected in each sample. Honey bee colony population size was used as a proxy for colony health by counting the number of frames more than 2/3 covered with bees (i.e. weak < 7 frames, average = 7–12 frames, strong >12 frames covered with bees). Included in the table are the mean number of pathogens per colony strength rating, the standard error estimate of the mean, and the number of colonies per colony health rating within this cohort. This sample cohort was comprised of 52.2% strong, 37% average, 8.6% weak, and 2.2% dead colonies.
(PDF)
Honey bee colonies from two Minnesota-based commercial beekeeping operations that transport honey bee colonies to California for almond pollination were monitored for two years from January 2014 to January 2015. At the onset of the study in January 2014, beekeepers identified 12–16 queen-right colonies that successfully overwintered (n = 28), of which two died throughout the course of the study and were replaced by three novel colonies bringing the total unique colonies up to 31 throughout the course of the study (n = 31); repeated sampling of colonies resulted in 93 total samples. Honey bee colony population size was used as a proxy for colony health by counting the number of frames more than 2/3 covered with bees (i.e. weak < 7 frames, average = 7–12 frames, strong >12 frames covered with bees). Pathogen diagnostics was performed by PCR (1 = positive detection, 0 = not detected, NA—not assessed or no sample) and quantitative PCR (qPCR). Relative RNA equivalents were determined by qPCR and were natural log transformed for statistical analyses.
(XLS)
Pathogen prevalence varied significantly by sampling event. Honey bee samples were obtained from two Minnesota based, commercially managed honey bee colonies, twice when colonies were located in California before and immediately after almond pollination and thus reflect the pathogen prevalence during the almond bloom, and three times after the almond pollination event when colonies were located in Minnesota. Pathogen prevalence was measured by totaling the number of pathogens detected via PCR in each sample. Included are the average number of pathogens per sampling event, the standard error estimate of the mean, and the number of colonies per colony health rating within this cohort.
(PDF)
Mean pathogen prevalence is higher in samples from strong rated colonies, than in samples from average rated colonies. The number of pathogens detected by PCR, and mites (when infestation percentage surpassed 3 mites per 100 bees) were summed for each sample and averaged (y-axis) for each colony health rating (x-axis). Colony health was rated using colony size as a proxy, where colonies with <7 frames covered with bees were rated weak (n = 8, blue), 7–12 frames were rated average (n = 34), >12 frames were rated strong (n = 48, gold), or 0 frames were rated dead (n = 2, black). Points represent raw data, and center bars represent the mean colony pathogen prevalence, which are surrounded by the upper and lower standard error estimates. Significant differences between mean pathogen prevalence of colony health ratings was calculated using Tukey’s HSD post-hoc test (p-value < 0.05).
(EPS)
Colonies acquired the highest mean number of pathogens by the first sampling event after almond pollination. The number of pathogens detected by PCR, and mites (when infestation percentage surpassed 3 mites per 100 bees) were summed in each sample and averaged (y-axis) for each sampling event (x-axis). Sampling events were conducted once before (n = 28, dark brown), once during (n = 28, light brown), and three times after California almond pollination (n = 11 yellow, n = 14 orange, and n = 11 red). Points represent raw data, and center bars represent the mean pathogen prevalence, which are surrounded by the upper and lower standard error estimates. Significant differences between the mean colony-level pathogen prevalence between sampling events were calculated using Tukey’s HSD post-hoc test (p-value < 0.05). The mean pathogen prevalence was greater in sampling events carried out after almond pollination when the colonies were located in Minnesota (after 1, 2, and 3) compared to sampling events before and during almond pollination when the colonies were located in California (with the exception of the pairwise comparison between during and after 3).
(EPS)
Total pathogen abundance changed throughout the monitoring period, and was different between beekeepers. The natural log transformed values of individual pathogens measured by qPCR were summed (y-axis) and plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies rated dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
KBV abundance varied across the monitoring period in beekeeping operation 2, but was never detected in colonies from beekeeping operation 1. The natural log transformed values of the relative KBV abundance as determined by qPCR (y-axis) in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). KBV abundance in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on KBV abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
LSV2 abundance was weakly correlated with date of sampling, but was greater in dead colonies. The natural log transformed values of the relative LSV2 RNA abundance as determined by qPCR (y-axis in honey bee samples were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
LSV1 abundance changed throughout the monitoring period, and was different between beekeeping operations. The natural log transformed values of the relative LSV1 RNA abundance as determined by qPCR (y-axis in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
N. ceranae abundance changed throughout the monitoring period and was predicted to be greater in strong rated colonies, compared to weak rated colonies. The natural log transformed values of the relative N. ceranae abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
L. passim abundance was predicted to change throughout the monitoring period, but the abundance was greater in beekeeping Operation 1, than beekeeping Operation 2. The natural log transformed values of the relative L. passim abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). L. passim abundance in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on L. passim abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
BQCV abundance changed throughout the monitoring period, and levels of abundance were higher in beekeeping Operation 2, than in beekeeping Operation 1. The natural log transformed values of the relative BQCV abundance as determined by qPCR in honey bee samples (y-axis) were plotted by the date of sample collection (i.e. day of year, x-axis). Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). BQCV abundances in colonies were plotted separately by beekeeping operation to indicate the effect of beekeeper on BQCV abundance. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph and illustrate changes in the pathogen abundance individual colonies throughout the sampling period.
(EPS)
The residuals of the change in frame count (used as a proxy for changes in colony health rating), after accounting for the sampling date (i.e., “day of year”) (y-axis) were plotted relative to the natural log transformed values of the relative pathogen RNA abundance (x-axis) for (A.) DWV (B.) LSV1, (C.) LSV2, (D.) BQCV, (E.) KBV, (F.) N. ceranae, and (G.) L. passim. Live honey bee samples were collected from colonies with varying colony health ratings (i.e., dead (black diamonds), weak (blue triangles), average (green circle), and strong (yellow square)). The best-fit line (blue) surrounded by upper and lower standard error estimates (gray) indicates trends in the changes in frame count relative to pathogen abundance. After accounting for changes in frame counts over time, the only significant result was a negative relationship between KBV abundance and changes in frame count. Unique colony identifier numbers, with the first digit identifying the pallet and the second digit identifying individual colony, label each point on the graph.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.