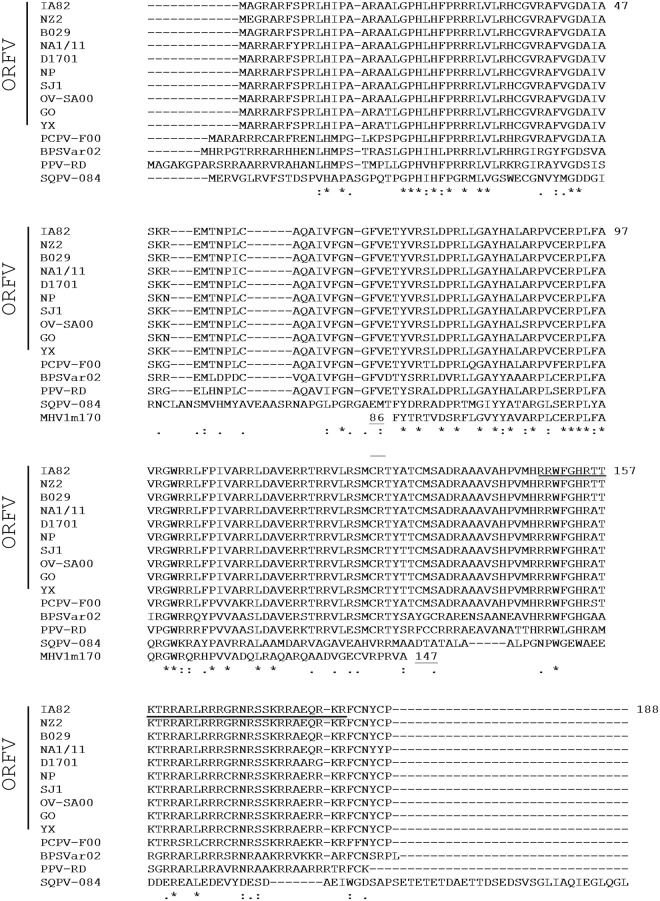
Fig 1. Clustal W alignment of amino acid sequences corresponding to parapoxvirus ORFV073 homologs, squirrellpox virus (SQPV)-084 protein, and murid betaherpesvirus m170 protein.
The ORFV strain IA82 sequence is at the top of the alignment, with amino acid positions indicated by numbers on the right. The predicted nuclear localization signal is shown underlined. Sequences listed below strain IA82 are arranged in order of decreasing % amino acid identity relative to strain IA82. For clarity, only the murid betaherpesvirus m170 domain with homology to parapoxvirus ORFV073 proteins is shown (m170 positions 86–147). Asterisks [*], colons [:], and periods [.] below the alignment indicate fully, strongly, and weakly conserved, residues, respectively. Viruses, strains, and GenBank accession numbers (in parentheses) used for the alignment are as follows: ORFV strains OV-IA82 (AY386263.1), NZ2 (DQ184476.1), B029 (KF837136.1), NA1/11 (KF234407.1), D1701 (HM133903.1), NP (KP010355.1), SJ1 (KP010356.1), OV-SA00 (NC 005336.1), GO (KP010354.1), and YX (KP010353.1); Pseudocowpox virus strain F00.120R [PCPV-F00] (GQ329669.1); Bovine papular stomatitis virus strain BV-AR02 [BPSVar02] (NC 005337.1); Parapoxvirus of the red deer in New Zealand strain HL953 [PPV-RD] (NC 025963.1); Squirrellpox virus strain Red squirrel UK [SQPV-084] (YP_008658509); Murid betaherpesvirus 1 strain N1 [MHV1m170] (CCE57166).