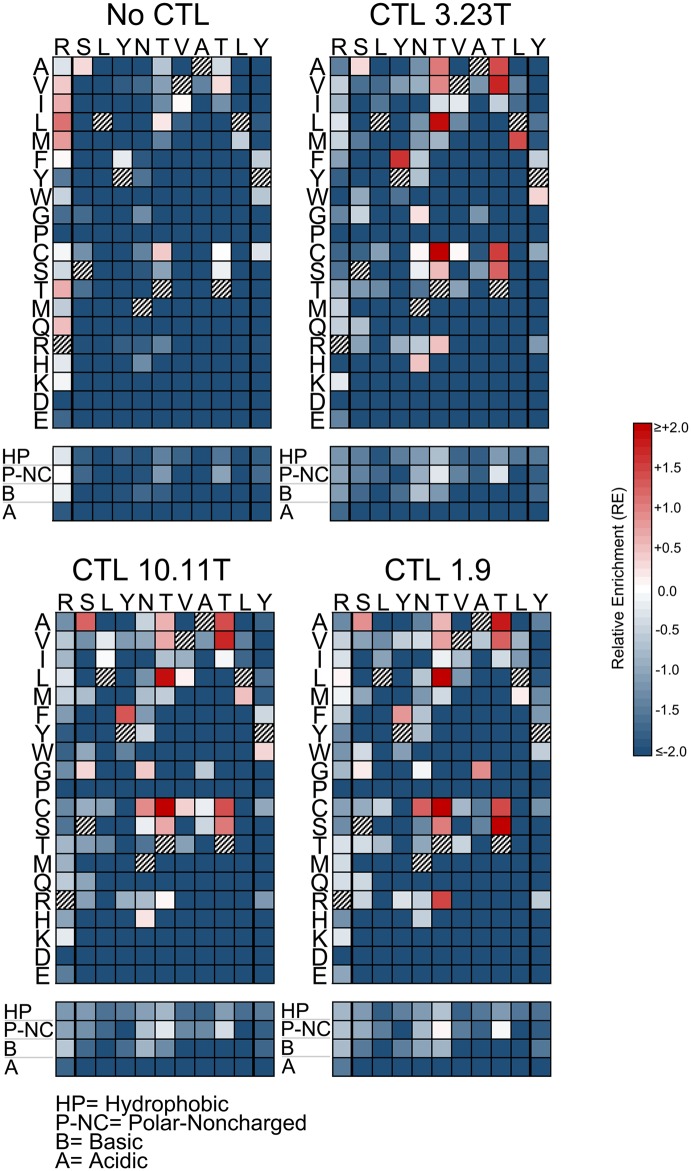
Fig 3. Single amino acid epitope variants display distinct patterns of selection in the absence or presence of CTLs (SL9 epitope).
The RE values of all single amino acid variants with or without addition of the indicated CTL clones are displayed as color-scaled boxes. The horizontal axis indicates each subtype B consensus amino acid of each epitope and its immediately flanking amino acids, and the vertical axis indicates substituting amino acids. Hatched boxes indicate consensus amino acids. Additionally, the mean REs for substitutions of amino acids that are hydrophobic (A, V, I, L, M, F, Y, W, G, and P), polar-noncharged (C, S, T, N, and Q), basic (R, H, K), or acidic (D and E) are indicated below each plot. Variants that were detected above threshold in the plasmid library but not in the virus library were considered non-replicating and assigned RE-CTL = -2.0 for these analyses.