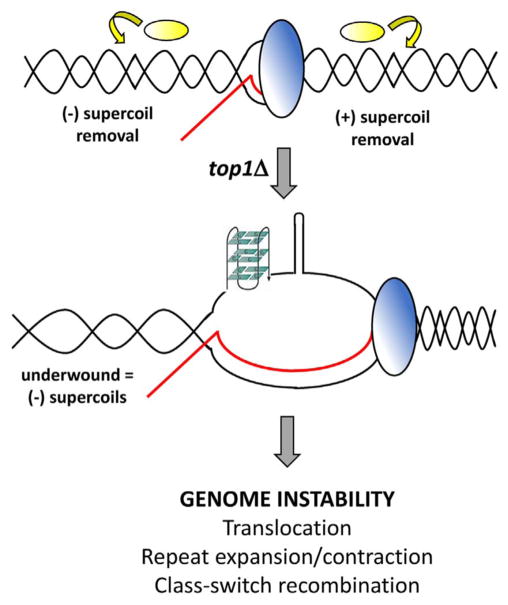
Fig. 1.
Genome stabilization by Top1 during transcription. During normal transcription by RNAP (blue oval), topological homeostasis is maintained by the activity of Top1 (yellow oval). In the absence of Top1, underwound and negatively supercoiled DNA that accumulates behind RNAP supports the formation of co-transcriptional R-loops in which the RNA transcript (red) pairs extensively with the DNA (black) template strand, and the non-template DNA strand is single-stranded. Single-stranded DNA folds into non-B secondary structures such as G4 DNA and hairpins. R-loops and non-B structures initiate genome instability.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)