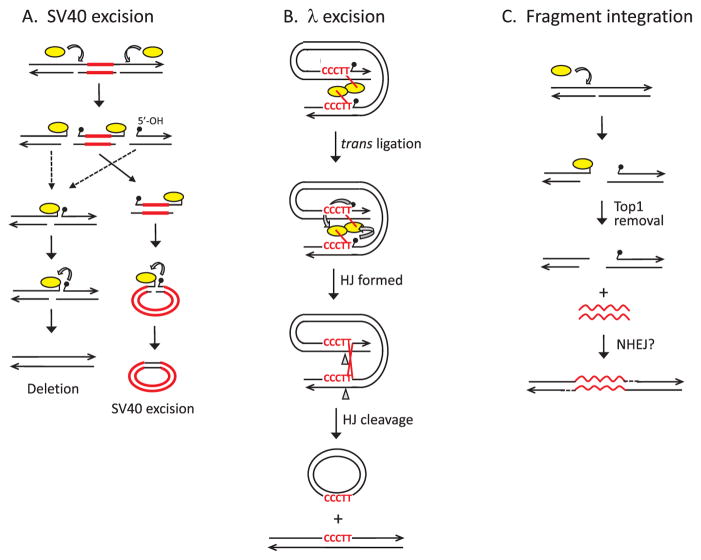
Fig. 2.
Top1-mediated genome instability. (A) Top1 (yellow oval) catalyzes excision of SV40 (red lines) from the genome. Cleavage of Top1 opposite a nick generates a DSB with the enzyme trapped at the ends. The relevant cleavages occur on the same DNA strand so that the excised fragment contains a Top1cc at one end and a 5′-OH (small black popsicle) at the other. Attack of the Top1-DNA linkage by the 5′-OH produces a nicked circular molecule, and the similar reaction can seal the broken chromosome. (B) Excision of λ from the E. coli genome by vcTop1. vcTop1 cleaves at 5′-CCCTT consensus sites that flank the λ genome. The vcTop1 complexes interact to facilitate trans ligation in which the 5′-OH generated by one enzyme attacks the phosphotyrosyl bond of the other. This creates a Holliday junction (HJ) that is resolved into crossover products by nicking and ligating the complementary, 5′-AAGGG-containing strands. (C) Top1 cleaves opposite a nick, generating a genomic DSB. Exogenous DNA (wavy red lines) is then joined to the broken ends by the NHEJ pathway or by Top1-mediated ligation (not shown). Black lines correspond to single DNA strands, with arrowheads indicating 3′ ends.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)