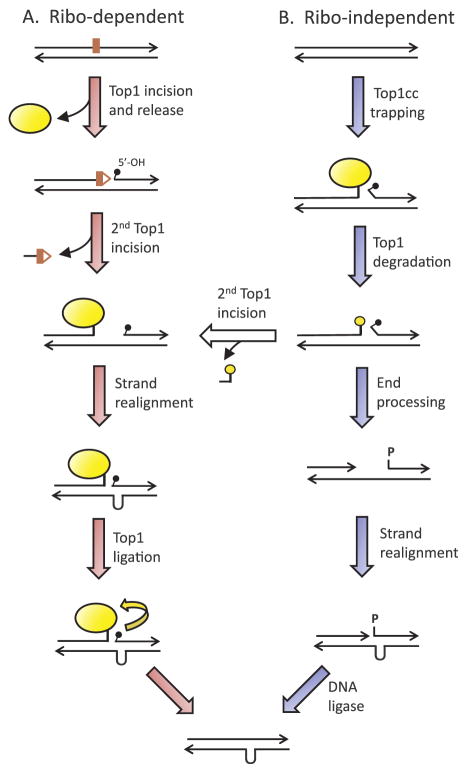
Fig. 3.
Mechanisms of Top1-dependent short deletions. (A) Ribo-dependent deletions are initiated when Top1 (yellow oval) cleaves at a ribo and is released from the DNA, leaving 2′,3′-cyclic phosphate (red triangle) and 5′-OH (small black popsicle) ends. A second Top1 cleavage upstream releases a small oligonucleotide and traps the enzyme. Repeat-mediated realignment of complementary strands brings the 5′-OH made by the first cleavage into the correct alignment for efficient enzyme-mediated ligation. (B) Ribo-independent deletions initiate with stabilization of a Top1cc. Following degradation of Top1, a second enzyme cleaves upstream and deletion formation proceeds as for ribo-dependent deletions. Alternatively, the ends can be processed into a gap flanked by a 3′-OH and 5′-phosphate. Realignment between complementary strands converts the gap to a nick that is sealed by ligase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)