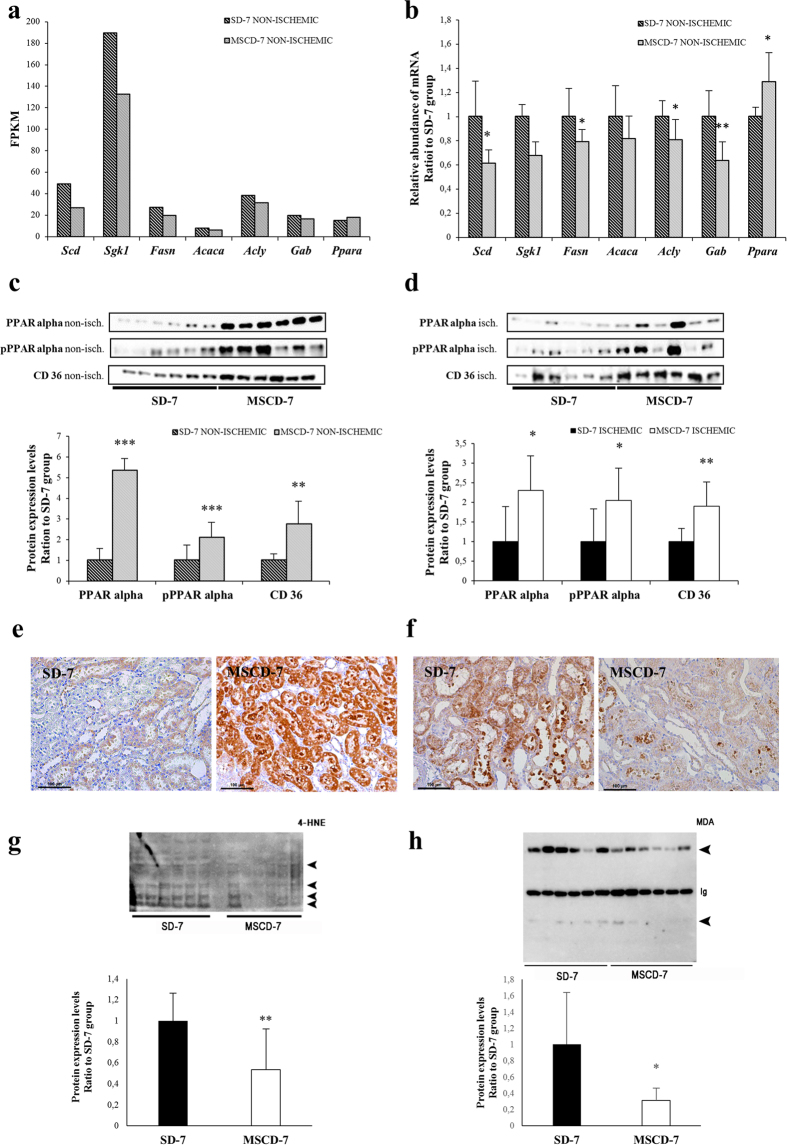
Figure 4.
Impact of MSC on lipid metabolism. (a) Significantly differentially expressed genes involved in fatty acid biosynthesis and nuclear receptor in lipid metabolism pathways in non-ischemic kidneys exposed to MSC (MSCD − 7, n = 6) versus saline (SD − 7, n = 6) 7 days before renal I/R, on the basis of the high-throughput RNA-sequencing. Data are shown in Fragments Per KiloBase per Million of mapped reads value. (b) RT-qPCR analysis of the genes corresponding to panel (a). The mRNA expression levels were standardized using GAPDH as housekeeping gene. (c,d) Quantification of PPARα, phospho-PPARα and CD36 expression in non-ischemic (panel c) and ischemic (panel d) kidneys. (e) Immunohistochemistry for FAT/CD36 in non-ischemic kidneys. (f–h) Representative immunochemistry of malondialdehyde (MDA) in renal parenchyma and quantitative immunoblotting of 4-hydroxy-2-nonenal (4-HNE) and MDA modified proteins (arrowheads) in ischemic kidneys of SD − 7 and MSCD − 7 groups (Ig, Immunoglobulins). Data are presented as mean ± standard deviation. Significant differences are indicated, *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001 versus appropriate control group.