Figure 1.
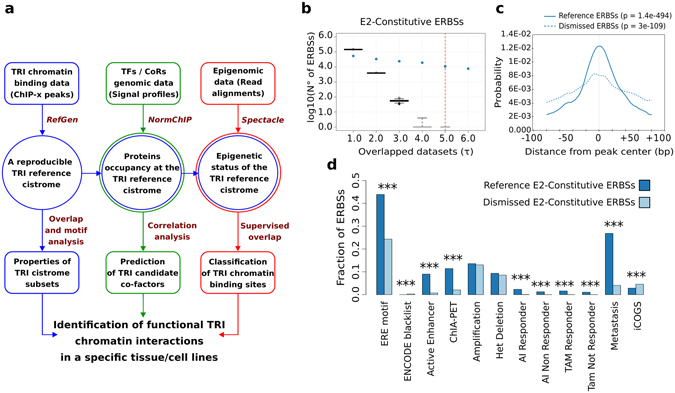
An integrative strategy to analyze ChIP data. (a) Schematic representation of our integrative strategy applied in the analysis of the cistrome of a Transcriptional Regulator of Interested (TRI). Each column represents an analytical step designed to characterize the reference cistrome (left column, blue boxes), the TRI candidate cofactors (center column, green boxes), and the epigenetic classes of TRI binding sites (right column, red boxes). Rectangles indicate input and output data and the main analytical methods applied are reported. TF, Transcription Factor; CoR, Co-Regulator. (b) Box plot representing as blue dots the number of ERα Binding Sites (ERBSs) identified in at least a specified number of ERα ChIP studies performed in MCF-7 grown in estrogens-enriched medium (E2-Constitutive experimental context). Black box plots represent the number of random genomic regions with the same length that are overlapped using the same threshold (τ) selected for the ERBSs analysis. The red dashed bar indicates the threshold corresponding to the 75% of studies that we selected to define the ERα cistrome for the E2-Constitutive experimental context. (c) Line plot representing the probability of the Estrogen Response Element (ERE) motif within a window of +/−100 bp centered on E2-Constitutive ERBSs belonging to the E2-Constitutive cistrome (Reference ERBSs) or that were filtered-out by our analysis (Dismissed ERBSs). At top, the p-value from the motif analysis is reported. (d) Fraction of E2-Constitutive ERBSs overlapping independent genomic features including: ERE motif, ENCODE blacklisted genomic regions, ERα bound active enhancers previously identified in MCF-7 (Active Enhancer), genomic regions of ERα-mediated long-range chromatin interactions (ChIA-PET), genomic regions amplified or heterozygous deleted in MCF-7, ERBSs identified in breast cancer tissue from patients before receiving Aromatase Inhibitor (AI) or Tamoxifen (Tam) and that responded (R) or not (NR) to treatment, ERBSs identified in distal breast cancer metastases, and the list of variants from the iCOGS project.
