Figure 1.
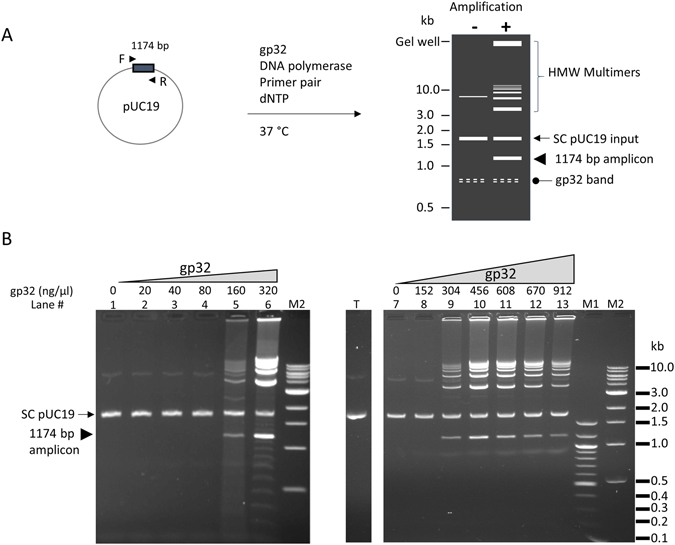
T4 gp32 protein-assisted isothermal amplification on circular dsDNA template pUC19. (A) schematic representation of the amplification reaction and product pattern after agarose gel electrophoresis. Primer pair consisting of the F and R primers (arrow heads) allows the amplification of a 1174 bp amplicon (box) of the pUC19 plasmid (circle). When there is no amplification, pUC19 migrates as a supercoiled band (SC pUC19, arrow) and weaker upper band. There is usually a faint and often diffuse band migrating at approximately 0.8 kb (oval arrow) whenever gp32 protein is present in the reaction. The amplification products consist of a band of the single amplicon (arrow head), high molecular weight (HMW) multimers (bracket) including those forming a ladder pattern and those retained in the gel well. HMW multimers migrate slower than the SC pUC19 input (arrow) and the linearized pUC19 (not shown). (B) Effect of gp32 concentration on the amplification of 0.1 μg of pUC19 template using primer #21: Left panel (lanes 1–6), low range of gp32 (20–320 ng/μl); Right panel (lanes 7–13), high range of gp32 (152–912 ng/μl). Lanes 1 and 7 contained no gp32 as negative controls. The location of the single amplicon (1174 bp) flanked by primer pair #21 is marked by an arrow head in the left panel. Lane T has 0.1 μg pUC19 alone without any treatment. M1 and M2 are 100 bp ladder and 1 kb ladder DNA markers respectively.
